the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Identification of new microbial functional standards for soil quality assessment
Michael Schloter
Berndt-Michael Wilke
Lee A. Beaudette
Fabrice Martin-Laurent
Nathalie Cheviron
Christian Mougin
Jörg Römbke
The activity of microorganisms in soil is important for a robust functioning of soil and related ecosystem services. Hence, there is a necessity to identify the composition, diversity, and function of the soil microbiome in order to determine its natural properties, functioning, and operating range as well as to assess ecotoxicological effects due to anthropogenic activities. Numerous microbiological methods currently exist in the literature and new, more advanced methods continue to be developed; however, only a limited number of these methods are standardised. Consequently, there is a need to identify the most promising non-standardised methods for assessing soil quality and to transform them into standards. In agreement with the “Ecosystem Service Approach”, new methods should focus more on soil microbial functions, including nutrient cycling and greenhouse gas emission, pest control and plant growth promotion, carbon cycling and sequestration, as well as soil structure development and filter function. The few existing standardised methods available that focus on the function of the soil microbiome mostly include measurements, like basal respiration, enzyme activities, and biodegradation of organic matter, under well-defined conditions in the lab. This paper sets out to summarise and expand on recent discussions within the International Organization for Standardization (ISO), Soil Quality – Biological Characterization sub-committee (ISO TC 190/SC 4), where a need was identified to develop scientifically sound methods which would best fulfil the practical needs of future users for assessing soil quality, going beyond the existing test systems. Of particular note is the current evolution of molecular methods in microbial ecology that use quantitative real-time PCR (qPCR) to produce a large number of new functional endpoints which are more sensitive as compared to “classical” methods. Quantitative PCR assesses the abundance of microbes that catalyse major transformation steps in nitrogen and phosphorus cycling, greenhouse gas emissions, chemical transformations including pesticide degradation, and plant growth promotion pathways based on the assessment of marker gene sequences that drive the related processes. In the assessment of soil quality methods, it was found that most methods focus on bacteria and related endpoints. Techniques to describe fungal communities as well as their functional traits are far less represented. As such, techniques to analyse fungal enzyme activities are proposed. Additionally, methods for the determination of microbial growth rates and efficiencies, including the use of glomalin as a biochemical marker for soil aggregation, are discussed. Furthermore, field methods indicative of carbon turnover, including the litter bag test and a modification to the tea bag test, are presented. However, it is obvious that with increasing developments in high throughput sequencing technologies and big data analyses, including metagenomics analysis, it will be possible to implement these technologies into the standardisation process for assessing the functions of the soil microbiome. Overall, it is suggested that endpoints should represent a potential function of soil microorganisms rather than actual activity levels, as the latter can largely be dependent on short-term variable soil properties such as pedoclimatic conditions, nutrient availability, and anthropogenic soil cultivation activities.
- Article
(376 KB) - Full-text XML
- BibTeX
- EndNote
Soils are one of the world's hotspots for biodiversity (Parker, 2010). Biota – both micro- and macro-organisms – in soil form strong networks and complex food webs, which determine the efficacy of the soil ecosystem functions (e.g. nutrient cycling, C storage and turnover, water retention, and modulation of soil structure) (Creamer et al., 2016). These functions support a range of ecosystem services that are indispensable for soil use in agriculture, horticulture, or silviculture (Nannipieri et al., 2017). At the same time, soil biota are strongly impacted by various anthropogenic activities, including ongoing global and climate change, pollution, as well as degradation and destruction of the terrestrial environment (Gomiero, 2016; Montgomery, 2008; Wagg et al., 2014). Consequently, investigations of the soil biome structure and function became an emerging topic in soil and environmental sciences (Griffiths and Philippot, 2013). As such, the number of studies describing the ecology of soils and ecosystem functioning has increased significantly over the past few decades and has resulted in the development of new methods for the assessment of microbial communities (e.g. Guillaume et al., 2016; Tian et al., 2018). In comparison, the ecotoxicological assessment of human impacts (e.g. chemical pollution and mechanical compaction) using single-species tests, which are well-established methods, has remained constant (Brookes, 1995; Joergensen and Emmerling, 2006).
Characterising the natural state of a soil's biome is a quite challenging task. In addition to its huge structural and functional diversity, soil biomes are influenced by strong temporal dynamics including seasonal weather conditions and the enormous spatial heterogeneity, which ranges from field scale to microscale (Kuffner et al., 2012; Regan et al., 2014; Suriyavirun et al., 2019). All of these intrinsic properties hinder the interpretation of data obtained from the analysis of soil biomes and the measurement of their functional traits.
Despite the fundamental methodological advances over the past years, which allow for an in-depth analysis of microbiomes and, to some extent, other soil-living organisms (e.g. Joergensen and Emmerling, 2006; Paul, 2015; Yates et al., 2016), only a limited number of soil biological methods have been standardised (for details, see Sect. 3). As a result, large and often significant deviations are observed in the results obtained when non-standardised methods are used (e.g. Strickland and Rousk, 2010). This is especially true for methods that are based on high throughput sequencing approaches, where variability and bias in data can occur from the “wet-lab” steps right through the various bioinformatics pipeline analysis steps (Quince et al., 2017). Therefore, the comparability between datasets generated by different laboratories using different methods or modified protocols of the same method is problematic. Consequently, the development of quality indices and threshold values, respectively, for assessing soil quality is nearly impossible (Bastida et al., 2008). Presumably, this is why the number of meta-analyses in soil biology remains small.
Given that there is a lack of harmonisation between existing methods and, at the same time, a proliferation of new methods, there is a need to identify the most promising methods described in the literature that can be standardised to produce reliable indicators for soil quality (e.g. Philippot et al., 2012). At the Annual International Organization for Standardization (ISO) meeting of TC 190 (Soil Quality) in Fukuoka, Japan, in October 2013, a decision was made to compile a list of available methods and to identify those that would be suitable for assessing soil quality. Additionally, during a subsequent meeting of ISO TC 190/SC 4/WG 4 (Microbiological Methods) held in Paris, France, in March 2014, further discussions focused on the criteria for suitable methods to be comprised of microbial functional indicators. In this paper, we summarise the major outcomes of the discussions which took place over the past several years within ISO TC 190/SC 4. Therefore, besides collating a list of criteria for the selection of test methods for the future analysis of microbial functions in soil, the aim of this paper is to present our opinion, as members of the ISO TC 190 committee, to initiate further discussion on possible methods that should be standardised for future soil quality assessments.
Several papers addressing the task of identifying suitable methods to be used as biotic indicators (usually including faunal indicators) were published in the last few years, mainly in the context of EU research projects (e.g. Bispo et al., 2009; Faber et al., 2013; Ritz et al., 2009; Römbke et al., 2010). Here, we propose to base the selection of soil quality methods more on the “Ecosystem Service Approach” (MEA, 2005) which is increasingly recognised by both environmental scientists and regulatory agencies (Breure et al., 2012; Galic et al., 2012) and which takes into account that soils have been raised to the rank of a natural resource to be protected. As a consequence, and in addition to method development and application (including the assessment of biodiversity as a prerequisite for soil function), the focus of future activities should be the determination of soil microbial functions as recommended endpoints (Kvas et al., 2017; Nienstedt et al., 2012; van der Putten et al., 2010; TEEB, 2010). Consequently, we propose to assess both existing and new methods for the selection of microbial functional tests that support various soil ecosystem services. This structures our approach, simplifies the identification of ecologically relevant methods, and presumably increases their acceptance by users, including the regulatory and stakeholder community. The following soil functions and ecosystem services have been defined and are proposed to be used as a starting point for the development of future methods (MEA, 2005; Ockleford et al., 2017):
-
biodiversity, genetic resources, cultural services;
-
food web support;
-
biodegradation of pollutants;
-
nutrient cycling (for example, N and P);
-
pest control and plant growth promotion;
-
carbon cycling and sequestration;
-
greenhouse gas emissions; and
-
soil structure affecting soil water, gas balance, and filtration function.
A second major criterion for selecting methods for standardisation is their usability. The method should be applicable in regulations (e.g. European and national agencies registering chemicals or products) and for the evaluation of soil ecology and functioning as fundamental aspects of soil quality (e.g. by stakeholders and researchers). Moreover, the routine use of methods to inform farmers and site owners on soil quality as continuous assessments of their land and land-use practices could be an additional condition that would require the choice of easy-to-use methods or possibly encourage the simplification of existing methods. Overall frequently used methods generate more data, which in turn is of high importance for the validation of threshold values. Therefore, the aim of this process is to identify methods that are scientifically sound and best fulfil the practical needs of future users. The most appropriate new methods, including those proposed in this article, need to be evaluated using the criteria required for the standardisation of ISO methods.
To assess possible methods, a list of criteria was used based on the “logical sieve” approach (Ritz et al., 2009). The list of criteria for the identification of functional indicators and associated methodologies (Table 1) was an outcome of the EU FP7 EcoFINDERS project (Faber et al., 2013). The criteria were compiled after sending a questionnaire to 25 partner institutions primarily working in the field of environmental science, mainly representing academia but also regulators and subcontracting laboratories. These criteria are applicable for different kinds of indicators and methods, including those addressing the functions of soil microbial communities. In the following sections, we assume that existing ISO standardised methods partly already fulfil these criteria, but not all relevant endpoints can be measured.
Table 1List of criteria for the selection of indicators for microbial functional indicators, based on Faber et al. (2013) and Pulleman et al. (2012), with slight modifications by the authors.
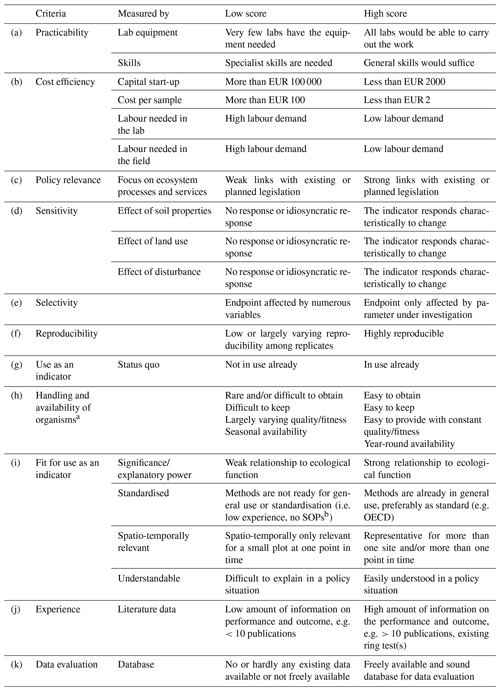
a Only relevant for faunal species. Does not apply to soil microorganisms that are tested with their natural abundance in mixed communities. b Standard operating procedures.
Current methods that have already been implemented as ISO standards are found in Table 2, whereas methods that might be considered for future standardisation are in Table 3. The compilation in Table 2 comprises methods to quantify microbial biomass (e.g. through fumigation extraction of microbial biomass carbon (MBC) and DNA) (Function 6) as well as for (further) analysis of structural microbial diversity (e.g. determination of microbial fingerprints by phospholipid fatty acids (PLFA) analysis) (Function 1). Additionally, microbial biomass, measured as respiratory activity, has been included in Table 2 but is not directly linked to one of the ecosystem services, as it provides important information on the activity of the complete microbiome (i.e. microflora and microfauna). Soil basal respiration normalised to MBC (ISO 14240-1, 1997; ISO 14240-2, 1997, Table 2) yields the metabolic quotient qCO2, which is a sensitive indicator for microbial carbon use efficiency (Anderson and Domsch, 1993). However, its use as an endpoint to assess anthropogenic and natural impacts on the soil microbiome has been controversially discussed in the literature (Wardle and Ghani, 1995). The microbial quotient (MBC related to the organic carbon content of a soil) is an indicator revealing changes in the microbial dynamic equilibrium of soils in response to exposure to natural or anthropogenic stressors (Pankhurst et al., 2001).
Table 2Methods already validated and published as ISO standards for determining potential microbial biomass and activities for soil quality.
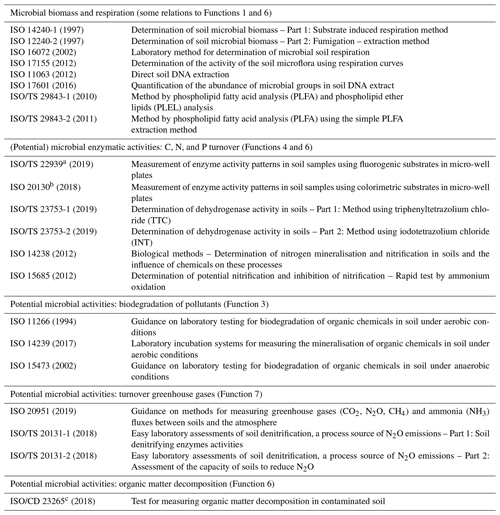
a Measured enzyme activities: arylsulfatase E.C. 3.1.6.1; α-glucosidase E.C. 3.2.1.20; β-glucosidase E.C. 3.2.1.21; β-xylosidase E.C. 3.2.1.37; cellobiosidase E.C. 3.2.1.91; N-acetylglucosaminidase E.C. 3.2.1.52; phosphodiesterase E.C. 3.1.4.1; phosphomonoesterase E.C. 3.1.3.2; leucine-aminopeptidase E.C. 3.4.11.1; alanine-aminopeptidase E.C. 3.4.11.12. b Measured enzyme activities: Arylamidase E.C. 3.4.11.2; arylsulfatase E.C. 3.1.6.1; α-glucosidase E.C. 3.2.1.20; β-glucosidase E.C. 3.2.1.21; β-galactosidase E.C. 3.2.1.22; N-acetylglucosaminidase E.C. 3.2.1.52; phosphatase E.C. 3.1.4.1; acid phosphatase E.C. 3.1.4.1; alkaline phosphatase E.C. 3.1.4.1; urease E.C. 3.5.1.5. c Degradation of cellulose under laboratory conditions.
The biodiversity function (Function 1) addresses parameters related to the structural diversity of the soil microbiome. Here, respective ISO guidelines analysing PLFA, phospholipid ether lipids (PLEL) (ISO/TS 29843-1, 2010; ISO/TS 29843-2, 2011), and DNA (ISO 11063, 2012; ISO 17601, 2016) have already been well implemented into guidelines (Table 2). In addition, high throughput sequencing of barcodes of the ribosomal operon (16S rRNA gene for bacteria and archaea and ITS (internal transcribed spacer) region for fungi) has generated a large amount of data (Schöler et al., 2017). These approaches have also been used successfully for other microbial groups like protists (using the 18S rRNA gene as a target). As it is well accepted that the use of different primer pairs introduces different biases (Ramirez et al., 2018), standards have been recommended by international initiatives. For example, the Earth Microbiome project (http://www.earthmicrobiome.org, last access: 27 January 2020) recommended a primer pair targeting the V4 region of the 16S rRNA gene and ITS2 region for bacterial and fungal barcoding, respectively. Bioinformatics pipelines used for barcoding approaches are already well standardised and shared worldwide among the scientists, which makes possible the cross-comparison of various datasets from different labs. Unfortunately, this is not yet the case for the pipelines to analyse metagenomics datasets that are still under constant evolution, making cross-comparisons difficult.
Although microbial diversity, per se, is not strongly correlated with a particular functional capacity, it is clear that the loss of diversity can have an impact on microbial function (Thiele-Bruhn et al., 2012), at least for relatively specific functions performed by narrow microbial guilds or taxa. This applies even more when certain taxa are closely linked to very specific functions, including nitrifiers, methanogens, and arbuscular- and ecto-mycorrhizal fungi and biocontrol microorganisms like Trichoderma (e.g. Hartmann et al., 2009; Hayat et al., 2010; Lugtenberg and Kamilova, 2009; Peng et al., 2008; Singh et al., 2007; Xia et al., 2011). Therefore, the interpretation of the outcomes from microbial community-based testing tends to be straightforward and closely linked to Function 4 and Function 5.
Food web support (Function 2) of higher trophic levels no doubt starts from soil microorganisms and propagates through the trophic levels (e.g. earthworms) that are consumed by birds and mammals (Haynes, 2014; Scheu et al., 2002, 2005). However, the role of the microbiota in the soil food web is not fully understood, since many eukaryotic organisms can be considered meta-organisms, which carry their “own microbiome” that itself is essential for life-supporting functions. From this, it is unclear whether environmental microbiomes and host-specific microbiomes complement one another. So far there have been no comprehensive methods (especially not those addressing microbial functions) or standards available to address this problem. The use of stable isotope labelling of select carbon or nutrient sources as a promising approach to follow food webs and degradation pathways might provide one possibility in the future to assess food webs in soil. This would be accomplished by combining carbon and nitrogen stable isotope flux determination with phylogenetic analysis of the microorganisms labelled with the stable isotopes (e.g. Coban et al., 2015; Traugott et al., 2013; Lueders et al., 2004).
Methods to assess the biodegradation of pollutants (Function 3), as described above, are already implemented into ISO guidelines (Table 2) and are part of legal frameworks including pesticide directives (EU Regulation1107/2009/EC; European Commission, 2009). A number of standard methods for the determination of the potential of soils to degrade organic chemicals (Function 3) under both aerobic (ISO 14239, 2017) and anaerobic (ISO 15473, 2002) conditions are available. However, in the past, the development of standard methods was mainly driven by the need to assess the ecotoxicological effects of anthropogenic activities, such as chemical contamination of soils, rather than to describe and understand the natural properties and functions of soils. Defining methods for the determination of adverse effects of contaminants on soil biota was not only done by ISO, but was also a major task of other organisations such as the Organization for Economic Co-Operation and Development (OECD). For example, there are OECD guidelines, test nos. 216 and 217, for testing the long-term effects of single-exposure chemicals on soil microbial nitrogen and carbon transformation, respectively (OECD, 2000a, b). As a result, it was decided early that the standardisation of methods for toxicity testing should not be the primary aim of the ISO sub-committee (ISO TC 190/SC 4). Metagenomics lead to the rapid discovery of new genes, which catalyse degradation processes of xenobiotics and, consequently, offer new insight into the study of ecology of microbial degraders. The transfer of this knowledge into operational bioindicators for the estimation of soil filtration capabilities (e.g. by using qPCR or RT-qPCR assays to quantify those genes and their expression in soils, respectively) will be one of the major tasks in soil science in the upcoming years. Jeffries et al. (2018) for example were able to successfully predict the degradation rates of organophosphorus-based pesticides in a contaminated soil by using metagenomics-based functional profiling.
Some of the existing standard methods that are listed in Table 2 focus on the estimation of enzyme activities useful for soil quality assessment, which mainly contribute to Function 4. Here, the potential dehydrogenase activity measurement is an indicator of general (potential) oxidoreductase activity in soil. Since this measurement has been frequently used, there are large amounts of baseline data available on the toxic effects of a range of pollutants in soil. Recently, additional potential enzyme activities related to the C, N, P, and S cycle have been used and are either standardised or are in the process of standardisation.
The current evolution of molecular methods in microbial ecology has resulted in a large number of new endpoints. It is well known that many of the new endpoints (e.g. using quantitative real-time PCR (qPCR)) are more sensitive than classical methods that had been standardised in the past (Ribbons et al., 2016; Schulz et al., 2016). This new metagenomics approach will be of high importance in the future, as it allows for the implementation of information on new functional traits that can be standardised into an analytical pipeline. Direct sequencing of soil DNA extracts, called metagenomics, is nowadays a method that can easily be implemented to obtain an overview of in-soil living organisms, including microorganisms. It is recognised that metagenomics is no longer limited by sequencing possibilities, but more by the availabilities of large-scale computing clusters to analyse the amount of data generated. The future success of metagenomics in soil surveys will mainly depend on the possibilities to standardise bioinformatics pipelines as well as on the availability of tools for big data analysis and artificial intelligence. However, it must be mentioned that even a well-standardised and automated workflow will generate only data on the relative abundance of nucleotide sequences and not absolute values (Geisen et al., 2019). Thus at the moment for the assessment of new methods linked to Functions 4 to 8, qPCR from soil DNA extracts (ISO 17601, 2016) plays a very important role in determining the abundance of single-marker gene sequences, which are indicative of specific transformation processes or soil functions. For example, the quantification of nitrogen-fixing microbes, nitrifiers, and denitrifiers has been successfully implemented using the nifH, amoA, and nirS/nirK genes as markers, respectively (Henry et al., 2004; Hirsch et al., 2010; Ollivier et al., 2010; Sessitsch et al., 2006). Similarly, the quantification of microorganisms involved in the β-ketoadipate pathway has been implemented by targeting pcaH (El Azhari et al., 2008) and catA (El Azhari et al., 2010) gene sequences. Various methods for the assessment of soil microbial Function 4 (nutrient cycling), Function 5 (pest control and plant growth promotion), and Function 7 (greenhouse gas emissions) are proposed based on the qPCR analysis of gene sequence coding for enzymes which trigger the respective function (e.g. Fish et al., 2013; Ribbons et al., 2016; Smith and Osborn, 2009). Additionally, it should be noted that molecular methods based on the assessment of specific marker genes for estimating the degradation potential in soil have already been proposed for both PAHs (e.g. Cebron et al., 2008) and individual pesticides (e.g. Martin-Laurent et al., 2004). These could be interesting for future standardisation; however, if a method is very compound-specific and targeted, this could limit its application range. Thus, these specific approaches will not be discussed further in this article.
Major advantages of qPCR assays to quantify gene sequence numbers, which can be used as proxies for a given microbial process, are that they are (i) highly standardised, sensitive, selective, and reproducible, (ii) designed for high throughput analysis, (iii) available for a wide range of targets, and (iv) methods that are relatively cheap once the necessary analytical devices are on hand. Some training on the method is required; however, once trained the assays are easy to perform. For example, numerous studies have already used the microbial functional genes involved in nitrogen cycling to determine the status and to assess induced changes in the soil microbial community (Levy-Booth et al., 2014; Nannipieri and Eldor, 2009; Wallenstein et al., 2006). Consequently, the number of functional genes that are suited for use as specific indicators of soil function are continuing to grow in the literature as researchers gain experience in this field and data become more prevalent. Also, evidence is increasing that functional gene abundance and community structure are closely linked to related microbial activities and their increase or decrease, e.g. through agricultural fertiliser regime or soil contamination (Levy-Booth et al., 2014; Ouyang et al., 2018; Xue et al., 2018). However, contrasting findings have also been reported, pointing to the fact that functional gene abundance and diversity is less affected by short-term changes, e.g. due to soil moisture changes (Zhang et al., 2019). A critical meta-analysis of existing data and reports, respectively, would be timely to better identify and generalise the linkage of functional gene abundance and ecosystem services.
Disadvantages, on the other hand, are that (i) the quality of qPCR data depends on soil DNA extracts (PCR inhibition), (ii) primer pairs, even degenerated ones, might not successfully amplify all microbes of the functional group of interest, (iii) only genetic potential is resolved, and (iv) there is no differentiation between active, dormant, or dead microorganisms, when working with DNA as a template for the qPCR reaction. The analysis of total RNA and of mRNA, which could help to overcome the latter problem, is currently not a suitable alternative as it is highly dynamic in time and space and needs special care to stabilise the RNA extracted from complex environmental matrices to avoid its degradation. Another problem of DNA analysis is that the biological representativeness of the results is solely based on a relatively small amount of soil (from a few hundred mg to tens of g of soil) from which the DNA is extracted. The use of small soil samples (<1 g) simplifies the sample preparation process for molecular biologists; however, it provides a poor representation of the indigenous soil microbial community in the naturally inhomogeneous soil. Typically, the α-biodiversity declines with sample size, while that of β-biodiversity increases (Nicol et al., 2003; Penton et al., 2016). Lastly, it must be noted that the high repeatability and reproducibility of molecular biology methods, including qPCR assays, depends on extraction, purification, and amplification of DNA or RNA. This is typically performed using commercial extraction kits; however, simply changing the commercial supplier of a kit can substantially change the results (Brooks et al., 2015; Feinstein et al., 2009). This clearly challenges standardisation since standard methods must not hinge on a specific supplier.
Recently, molecular tools for the assessment of the microbial phosphorous turnover (Function 4) have been published (Bergkemper et al., 2016) where metagenomics data have been used for the construction of primers for P mineralisation, transport, and uptake. As another example, the relevance of anaerobic ammonium oxidation (anammox) for N cycling in soils has increased (Levy-Booth et al., 2014) along with the development of analytical methods for high throughput analysis. Among the microorganisms in soil that substantially govern pest control and plant growth promotion (Function 5), arbuscular mycorrhizae and ectomycorrhizal fungi are of high relevance. These microorganisms are especially abundant in the rhizosphere (Hartmann et al., 2009; Hayat et al., 2010; Lugtenberg and Kamilova, 2009). Methods related to Function 5 are listed in Table 3.
Table 3Potential new methods for the ISO standardisation process and assessment according to the “logical sieve” selection criteria (described in Table 1).
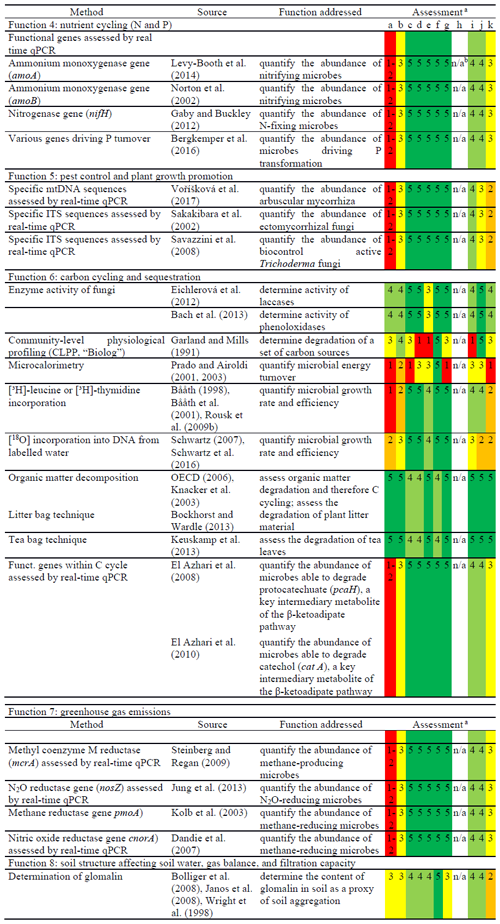
a Overall scoring in case of several measures for one criterion. Fulfilment of the criterion described by numbering (colour code): 1 (red) – very low; 2 (orange) – low; 3 (yellow) – medium; 4 (light green) – good; 5 (dark green) – very good. b n/a – not applicable.
Several options exist for (additional) standardised methods to test Function 6 (carbon cycling and sequestration) (Table 3). For Function 6, there is a need to implement more fungal activity analysis as most tests described mostly assess bacterial activities. Thus, the integration of more fungal enzyme activities into the suite of standardised methods for soil quality assessment is essential (for example, determining the turnover of complex natural compounds such as lignin) (Baldrian, 2006). The ligninolytic enzymes laccase and Mn-peroxidase, as well as the chitin-degrading 1,4-α-N-acetylglucosaminidase, are typical fungal enzymes of interest for ecosystem services (Jiang et al., 2014; Šnajdr et al., 2008). However, since other organisms also produce these enzymes, including bacteria and plants (Bollag, 1992; de Gonzalo et al., 2016), current methods do not specifically target fungal enzyme activities. As a result, the implementations of molecular methods for assessing fungal communities are far less developed than those for bacterial communities (Table 3). The first approaches to close this gap have been made in recent years. For example, molecular systems to detect gene coding for laccases and other ligninolytic enzymes as well as other fungal activities in carbon cycling have been published and applied (Edwards et al., 2011; Chen et al., 2013; Hannula and van Veen, 2016). Also for genes involved in the fungal nitrogen turnover, primer pairs have been successfully developed (Gorfer et al., 2011). However, here, even more than for bacteria, it is critical to link copy numbers of genes directly to the size of the respective functional population, as in many cases one organism can harbour multiple operon coding for the same genes involved in a given function.
The method of community-level physiological profiling (CLPP) using the Biolog™ system (Biolog, Hayward, CA, USA) was first developed in the late 1980s to identify bacteria of clinical importance by assessing the consumption of 95 different carbon sources in a microtiter plate. The technique was then extended to identify bacterial strains from environmental mixed microbial communities samples using select carbon sources (Garland, 1997). Currently, the technique is frequently used to assess the effects of contaminants on soil microbial activity (Bloem and Breure, 2003; Schmitt et al., 2004). As such, the CLPP method has become a measure of microbial functional diversity in soil (e.g. Gomez et al., 2006) and was used to distinguish the biodiversity of soil microbial communities in monitoring programs (Rutgers et al., 2016). Even though the method is easy to use, it does have some drawbacks (Winding and Hendriksen, 2007). The technique is based on the utilisation of select carbon sources, which when consumed result in reduction, and thus colour change, of a tetrazolium indicator dye (Garland and Mills, 1991). This reaction is based on the dehydrogenase enzyme activity of cultivable, fast-growing, aerobic, eutrophic microorganisms (largely bacteria). Consequently, this technique does not reflect the full spectrum of microbial species within a mixed soil community. Additionally, due to the artificial growth conditions required in the test, it is argued that the method does not reflect the microbial community diversity and its function of a given soil (Glimm et al., 1997). On the other hand, however, standardised conditions allow for direct comparisons between microbial communities in different sites, for example, independent of the abiotic conditions, thus making CLPP a popular method for toxicology testing (Preston-Mafham et al., 2002).
Isothermal micro-calorimetry is another technique that involves the direct measurement of energetics in soil and provides a functional link between energy flow and the composition of belowground microbial communities at a high taxonomic level (Herrmann et al., 2014). With this method, an integrative determination of the metabolic activity of soil bacteria and fungi is achieved. The integrated assessment of substances' and energy turnover has high potential to elucidate the regulation of soil ecological functions. However, the substantial costs for the acquisition of this very specific instrumentation are considered a major drawback. Furthermore, the measurement requires water saturation of the soil and, thus, the samples are modified. Since calorimetry has been rarely used and data and publications are few, this method is considered not ready for standardisation.
The methods targeting thymidine or leucine incorporation into microbial biomass can be used to determine microbial growth rates and efficiencies (Bååth et al., 2001; Rousk, 2016). Growth rate is a fundamental reference for numerous other microbial properties and functions. For example, it is required to calculate microbial carbon use efficiency (CUE) as a key parameter describing C-substrate turnover and storage in soil (Liu et al., 2018; Spohn et al., 2016; Takriti et al., 2018). Furthermore, the method can be used to assess the adverse effects of toxic chemicals on the microbial community (Modrzyński et al., 2016; Rousk et al., 2009a). The drawbacks of these two methods are that (i) specific training is required, (ii) laboratories must have a permit to manipulate radioactive isotopes, and (iii) there are higher costs for proper handling and disposal of 3H-labelled radioactive material. As an alternative, the incorporation of the stable isotope 18O from labelled water into soil microbial DNA can be used to distinguish growing and non-growing microorganisms based on the gradient separation of [18O]DNA and [16O]DNA (Schwartz, 2007). The 18O stable isotope method has been improved by sequencing a marker gene from fractions retrieved from ultracentrifugation to produce taxon density curves, thus enabling researchers to estimate the percent isotope composition of each microbial taxon's genome (Schwartz et al., 2016). This method continues to be advanced and, although not used often, could have a high potential for future standardisation.
There are simplistic methods available to determine organic matter decomposition, which are indicative of C cycling (Function 6). The tests listed in Table 3 are based on measuring the weight loss of introduced organic materials of different complexity in soil over time. The tests are relatively easy to perform and inexpensive; however, degradation activity is not exclusive to microorganisms, but can also include invertebrates. The OECD litter bag test (OECD, 2006) for site-specific assessment of organic matter decomposition uses wheat straw as the substrate and provides clear evidence of cellulose degradation. In general, the litter bag tests provide evidence of the degradation of naturally occurring plant material in soil. Results do, however, depend on the mesh size of the litter bags (increasing exclusion of soil animals with decreasing mesh size). On the other hand, plant material or litter is hard to standardise, with the results largely depending on the composition of the plant material. As such, artificial cellulose has been successfully used for a laboratory procedure to assess organic matter decomposition (Kvas et al., 2017). Another alternative to the litter bag test is the use of tea bags (Keuskamp et al., 2013). Tea bags can be purchased to contain a consistent quality of material, and so this method is preferred by citizen science (e.g. farmers to assess the soil quality of their land). In order to better distinguish the degrading abilities of different soil microbiomes, the test could be modified to use different types of tea that contain recalcitrant material to a different extent. Another test for future method development is the bait-lamina test (ISO 18311, 2016) used to assess the degradation of organic matter in field soil by grazing invertebrates (Jänsch et al., 2013; Kvas et al., 2017). It is a simple test that can easily be adapted for use under controlled laboratory conditions (Jänsch et al., 2017).
Methods for the determination and assessment of greenhouse gas emissions from soil (Function 7) have already been standardised or are well advanced in the standardisation process (Table 2). They are mostly focused on measuring concentrations of greenhouse gases, like CO2, CH4, and N2O, as well as their fluxes as endpoints. In addition, molecular biology methods that estimate the relative abundance of functional microbial guilds or taxa give new insight into the ecology of microorganisms involved in the formation of greenhouse gases. For example, the qPCR measurement of key N2O functional genes has allowed researchers to link N2O reduction capacity to reduced greenhouse gas emissions in soil amended with organic matter (Xu et al., 2018). Additionally, the quantification of functional gene sequences related to methane generation and methane oxidation, respectively, yields detailed insights into the functional potential of climate change-affected permafrost soils (Yergeau et al., 2010).
For Function 8 (soil structure affecting soil water, gas balance, and filtration function), there is clear evidence that microbial activity and biomolecules substantially contribute to the formation and stability of micro-aggregates and thus to the structure, pore system, and pre-consolidation stress of soils (Six et al., 2004). While existing parameters, such as enzyme activities, are not clearly indicative in this regard (Beck and Beck, 2000), glomalin can be considered a biochemical marker of soil aggregation. This glycoprotein is produced by microorganisms, especially arbuscular mycorrhiza fungi, and significantly increases aggregate formation and stability (Rillig, 2004; Rillig and Mummey, 2006). The existing protocols for extraction (chemical extraction combined with autoclaving) and determination of glomalin, either by using the Bradford protein assay, enzyme-linked immunosorbent assay (ELISA), or the LC-MS method (Bolliger et al., 2008; Janos et al., 2008), open the possibility for its standardisation in the near future. It should be noted, however, that a well-equipped and experienced laboratory is required to perform this method.
As recently underlined by the European Food Safety Agency (EFSA) in a scientific opinion “addressing the state of the science on risk assessment of plant protection products for in-soil organisms”, there is an urgent need to modernise pesticide risk assessment by implementing specific protection goals for in-soil organisms which are key drivers of a wide range of functions supporting ecosystem services (Ockleford et al., 2017). There currently exists a multitude of methods that can potentially be used for this task. Here, we have identified in the body of this paper a number of methods that are presumably suitable for further evaluation and standardisation with regard to their scientific value and practical applicability. These prospective standardised methods will not only be useful to identify adverse effects on the soil microbiome, but also to conduct comparable studies in laboratories all over the world to define normal operating ranges of microbial activity in soil and respective quality indices and threshold values.
It is clear that all parameters taken together reflect the potential of a microbial community to perform a certain function and not solely a specific (actual) activity. This is important to understand in order to interpret the values of a given endpoint in relation to both energy fluxes and compound transformation rates, which can largely depend on intrinsic properties such as pedoclimatic conditions, nutrient availability as well as extrinsic properties such as anthropogenic effects, and soil cultivation measures. To make use of these methods as indicators of soil quality, there are several requirements that need to be included. These involve the assessment of the normal operating ranges of soil that include natural and dynamic fluctuations of a given endpoint. The methods need to be implemented into a framework, which takes into account site-specific conditions including soil type, pedoclimate, and land use. Undoubtedly, this requires further joint efforts in order to generate comprehensive databases from which normal operating ranges of values for a given proxy can be read. Such a task calls for standardised methods to obtain comparable data. Additionally, there is a requirement for the assessment of resistance and resilience of a given microbial endpoint to see how much it is affected by a soil disturbance and whether or not it can recover (e.g. return to its original state) after the disturbance has disappeared. Here the use of DNA-based methods, which provide a measure of a microbial community's potential to perform a given process, might be more useful than using RNA. The RNA rather indicates actual activities, which may highly fluctuate in time and space and thus are of less significance as an indicator. However, free DNA released from dead microbes is often highly resistant in soil, which might result in an overestimation of a potential function. This needs to be taken into account when interpreting the data. Recently, methods that extract DNA only from living cells have been described, but their use has not been introduced yet into recent standardisation activities.
Also, the use of a test battery to measure a range of interconnected endpoints is recommended (Ockleford et al., 2017) to integrate different biological and other parameters (e.g. soil pH, organic carbon content) into multiparametric indices (Bastida et al., 2008; Kvas et al., 2017). At present, it appears to be favourable to use a suite of different methods, i.e. functional gene analysis and microbial enzyme and/or degradation activities, for soil quality testing. Finally, to fully understand soil microbial functioning, a task was envisioned to investigate the linkage between the genetic functional potential and the available resources, termed the soil metaphenome (Jansson and Hofmockel, 2018). This will require even further integration and assessment of multiple parameters and test methods. Reaching that goal will surely promote soil ecological research but, at the current stage, may clearly go beyond the applied aim of standardisation to release easy-to-use targeted methods.
The critical evaluation of existing and non-standardised methods is required to further select and standardise new methods to assess soil quality. For methods linked to the molecular analysis of soil microbiomes, there is a need to ensure that worldwide activities are synchronised to propose important standards that are well accepted by the scientific community. To improve the reproducibility of data it has been agreed that a complex mixture of microorganisms (MOCK) must be implemented as a control in every experiment, but so far no common agreement on the composition of a MOCK community has been reached. However, it is clear that if further developments of microbial bar coding and/or metagenomics methods are to be implemented into ISO guidelines, a MOCK is required.
ISO standardisation committees are open circles and the presented selection and valuation of methods may not be complete. Environmental scientists are solicited to propose new work items enlarging the current catalogue of biological methods for future standardisation. Accordingly, this opinion paper aims at initiating a broader discussion intended to improve the measurement of microbial functions for soil quality assessment. Lastly, it must be noted that standardisation of methods is inevitably a balancing act. On the one hand, standardisation provides defined methods that are essential for obtaining comparable data, e.g. for integration into large, joint databases. On the other hand, it requires setting a specific method for several years. Consequently, scientific progress cannot be easily adopted, or at least with a delay, considering that standards are revised every 5 years, which may be a barrier to the introduction of new approaches resulting from technological evolution, especially in the fast-developing field of molecular biology methods. Hence, it is also the aim of this paper to have an open discussion to identify the best suitable methods with an assumed longer period of validity.
No data sets were used in this article.
The text was jointly discussed and written by all the authors. A first version of the manuscript was drafted by JR, STB, MS, and BMW. Further detailed aspects were added during the preparation of the manuscript by FLM, NC, CM, and LAB. LAB also took care of English proofreading.
The authors declare that they have no conflict of interest.
The compilation of methods was developed during the meetings of the ISO TC 190/SC 4 working group 4 “Soil biological methods”. We thank our colleagues for all the fruitful discussions. We gratefully thank Lily Pereg for reviewing a first draft of this paper and we will keep her in our remembrance. Biochem-Env is a service of the investment d'Avenir infrastructure AnaEE-France, overseen by the French National Research Agency (ANR) (ANR-11-INBS-0001).
This paper was edited by Jeanette Whitaker and reviewed by Robert Griffiths and Emilia Hannula.
Anderson, T. H. and Domsch, K. H.: The metabolic quotient for CO2 (qCO2) as a specific activity parameter to assess the effects of environmental conditions, such as pH, on the microbial biomass of forest soils, Soil Biol. Biochem., 25, 393–395, https://doi.org/10.1016/0038-0717(93)90140-7, 1993.
Bååth, E.: Growth rates of bacterial communities in soils at varying pH: A comparison of the thymidine and leucine incorporation techniques, Microb. Ecol., 36, 316–327 https://doi.org/10.1007/s002489900118, 1998.
Bååth, E., Pettersson, M., and Söderberg, K. H.: Adaptation of a rapid and economical microcentrifugation method to measure thymidine and leucine incorporation by soil bacteria, Soil Biol. Biochem., 33, 1571–1574, https://doi.org/10.1016/S0038-0717(01)00073-6, 2001.
Bach, C. E., Warnock, D. D., Van Horn, D. J., Weintraub, M. N., Sinsabaugh, R. L., Allison, S. D., and German, D. P.: Measuring phenol oxidase and peroxidase activities with pyrogallol, l-DOPA, and ABTS: Effect of assay conditions and soil type, Soil Biol. Biochem., 67, 183–191, 2013.
Baldrian, P.: Fungal laccases – occurrence and properties, FEMS Microbiol. Rev., 30, 215–242, 2006.
Bastida, F., Zsolnay, A., Hernández, T., and García, C.: Past, present and future of soil quality indices: A biological perspective, Geoderma, 147, 159–171, https://doi.org/10.1016/j.geoderma.2008.08.007, 2008.
Bloem, J. and Breure, A. M.: Microbial indicators, in: Trace Metals and other Contaminants in the Environment, edited by: Markert, B. A., Breure, A. M., and Zechmeister, H. G., Vol. 6, Elsevier, Dordrecht, 259–282, 2003.
Beck, T. and Beck, R.: Bodenenzyme. in: Handbuch der Bodenkunde, edited by: Blume, H.-P., Felix-Hennigsen, P., Frede, H.-G., Guggenberger, G., Horn, R., and Stahr, K., Wiley-VCH, Weinheim, Germany, 244 pp., 2000.
Bergkemper, F., Kublik, S., Lang, F., Krüger, J., Vestergaard, G., Schloter, M., and Schulz, S.: Novel oligonucleotide primers reveal a high diversity of microbes which drive phosphorous turnover in soil, J. Microbiol. Meth., 125, 91–97, https://doi.org/10.1016/j.mimet.2016.04.011, 2016.
Bispo, A., Cluzeau, D., Creamer, R., Dombos, M., Graefe, U., Krogh, P. H., Sousa, J. P., Peres, G., Rutgers, M., Winding, A., and Römbke, J.: Indicators for monitoring soil biodiversity, Integr. Environ. Assess., 5, 717–719, https://doi.org/10.1897/IEAM-2009-064.1, 2009.
Bockhorst, S. and Wardle, D. A.: Microclimate within litter bags of different mesh size: Implications for the “arthropod effect” on litter decomposition, Soil Biol. Biochem., 58, 147–152, 2013.
Bollag, J. M.: Decontaminating soil with enzymes, Environ. Sci. Technol., 26, 1876–1881, 1992.
Bolliger, A., Nalla, A., Magid, J., de Neergaard, A., Dole Nalla, A., and Bøg-Hansen, T. C.: Re-examining the glomalin-purity of glomalin-related soil protein fractions through immunochemical, lectin-affinity and soil labelling experiments, Soil Biol. Biochem., 40, 887–893, https://doi.org/10.1016/j.soilbio.2007.10.019, 2008.
Breure, A. M., De Deyn, G. B., Dominati, E., Eglin, T., Hedlund, K., Van Orshoven, J., and Posthuma, L.: Ecosystem services: A useful concept for soil policy making!, Curr. Opin. Env. Sust., 4, 578–585, 2012.
Brookes, P. C.: The use of microbial parameters in monitoring soil pollution by heavy metals, Biol. Fert. Soils, 19, 269–279, https://doi.org/10.1007/BF00336094, 1995.
Brooks, J. P., Edwards, D. J., Harwich, M. D., Jr., Rivera, M. C., Fettweis, J. M., Serrano, M. G., Reris, R. A., Sheth, N. U., Huang, B., Girerd, P., Strauss III, J. F., Jefferson, K. K., and Buck, G. A.: The truth about metagenomics: Quantifying and counteracting bias in 16S rRNA studies ecological and evolutionary microbiology, BMC Microbiol., 15, 66, https://doi.org/10.1186/s12866-015-0351-6, 2015.
Cebron, A., Norini, M. P., Beguiristain, T., and Leyval, C.: Real-time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHD alpha) genes from Gram positive and Gram-negative Bacteria in soil and sediment samples, J. Microbiol. Meth., 73, 148–159, 2008.
Chen, X., Su, Y., He, X., Liang Y., and Wu, J.: Comparative analysis of basidiomycetous laccase genes in forest soils reveals differences at the cDNA and DNA levels, Plant Soil, 366, 321–331, 2013.
Creamer, R. E., Hannula, S. E., Leeuwen, J., Stone, D., Rutgers, M., Schmelz, R. M., Ruiter, P., Hendriksen, N., Bolger, T., Bouffaud, M. L., Buee, M., Carvalho, F., Costa, D., Dirilgen, T., Francisco, R., Griffiths, B. S., Griffiths, R., Martin, F., Silva, P. D., Mendes, S., Morais, P. V., Pereira, C., Philippot, L., Plassart, P., Redecker, D., Römbke, J., Sousa, J. P., Wouterse, M., and Lemanceau, P.: Ecological network analysis reveals the inter-connection between soil biodiversity and ecosystem function as affected by land use across Europe, Appl. Soil Ecol., 97, 112–124, https://doi.org/10.1016/j.apsoil.2015.08.006, 2016.
Coban, H., Miltner, A., and Kästner, M.: Fate of fatty acids derived from biogas residues in arable soil, Soil Biol. Biochem., 91, 58–64, 2015.
Dandie, C. E., Miller, M. N., Burton, D. L., Zebarth, B. J., Trevors, J. T., and Goyer, C.: Nitric oxide reductase-targeted real-time PCR quantification of denitrifier populations in soil, Appl. Environ. Microb., 73, 4250–4258, 2007.
De Gonzalo, G., Colpa, D. I., Habib, M. H. M., and Fraaije, M. W.: Bacterial enzymes involved in lignin degradation, J. Biotechnol., 236, 110–119, 2016.
Edwards, I. P., Zak, D. R., Kellner, H., Eisenlord, S. D., and Pregitzer, K. S.: Simulated atmospheric N deposition alters fungal community composition and suppresses ligninolytic gene expression in a Northern Hardwood forest, PLoS ONE, 6, e20421, https://doi.org/10.1371/journal.pone.0020421, 2011.
Eichlerová, I., Šnajdr, J., and Baldrian, P.: Laccase activity in soils: Considerations for the measurement of enzyme activity, Chemosphere, 88, 1154–1160, https://doi.org/10.1016/j.chemosphere.2012.03.019, 2012.
El Azhari, N., Bru, D., Sarr, A., and Martin-Laurent, F.: Estimation of the density of the protocatechuate-degrading bacterial community in soil by real-time PCR, Eur. J. Soil Sci., 59, 665–673, 2008.
El Azhari, N., Devers-Lamrani, M., Chatagnier, G., Rouard, N., and Martin-Laurent, F.: Molecular analysis of the catechol-degrading bacterial community in a coal wasteland heavily contaminated with PAHs, J. Hazard. Mater., 177, 593–601, https://doi.org/10.1016/j.jhazmat.2009.12.074, 2010.
European Commission: EU-Regulation 1107/2009/EC of the European Parliament and the council of 21 October 2009 concerning the placing of plant protection products on the market and repealing Council Directives 79/117/EEC and 91/414/EEC, Offic. J. Europ. Union, L 309, 1–50, 2009.
Faber, J. H., Creamer, R. E., Mulder, C., Römbke, J., Rutgers, M., Sousa, J. P., Stone, D., and Griffiths, B. S.: The practicalities and pitfalls of establishing a policy-relevant and cost-effective soil biological monitoring scheme, Integr. Environ. Assess., 9, 276–284, 2013.
Feinstein, L. M., Woo, J. S., and Blackwood, C. B.: Assessment of bias associated with incomplete extraction of microbial DNA from soil, Appl. Environ. Microb., 75, 5428–5433, https://doi.org/10.1128/AEM.00120-09, 2009.
Fish, J. A., Chai, B., Wang, Q., Sun, Y., Brown, C. T., Tiedje, J. M., and Cole, J. R.: FunGene: The Functional Gene Pipeline and Repository, Front. Microbiol., 4, 291, https://doi.org/10.3389/fmicb.2013.00291, 2013.
Gaby, J. C. and Buckley, D. H.: A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase, PLoS ONE, 7, e93883, https://doi.org/10.1371/journal.pone.0093883, 2012.
Galic, N., Schmolke, A., Forbes, V., Baveco, H., and van den Brink, P. J.: The role of ecological models in linking ecological risk assessment to ecosystem services in agroecosystems, Sci. Total Environ., 415, 93–100, 2012.
Garland, J. L.: Analysis and interpretation of community-level physiological profiles in microbial ecology, FEMS Microbiol. Ecol., 24, 289–300, https://doi.org/10.1016/S0168-6496(97)00061-5, 1997.
Garland, J. L. and Mills, A. L.: Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization, Appl. Environ. Microb., 57, 2351–2359, 1991.
Geisen, S., Briones, M. J. I., Gan, H., Behan-Pelletier, V. M., Friman, V. P., de Groot, G. A., Hannula, S. E., Lindo, Z., Philippot, L., Tiunov, A. V., and Wall, D. H.: A methodological framework to embrace soil biodiversity, Soil Biol. Biochem., 136, 107536, https://doi.org/10.1016/j.soilbio.2019.107536, 2019.
Glimm, E., Heuer, H., Engelen, B., Smalla, K., and Backhaus, H.: Statistical comparisons of community catabolic profiles, J. Microbiol. Meth., 30, 71–80, https://doi.org/10.1016/S0167-7012(97)00046-8, 1997.
Gomez, E., Ferreras, L., and Toresani, S.: Soil bacterial functional diversity as influenced by organic amendment application, Bioresource Technol., 97, 1484–1489, https://doi.org/10.1016/j.biortech.2005.06.021, 2006.
Gomiero, T.: Soil degradation, land scarcity and food security: Reviewing a complex challenge, Sustainability, 8, 281, https://doi.org/10.3390/su8030281, 2016.
Gorfer, M., Blumhoff, M., Klaubauf, S., Urban, A., Inselsbacher, E., Bandian, D., Mitter, B., Sessitsch, A., Wanek, W., and Strauss, J.: Community profiling and gene expression of fungal assimilatory nitrate reductases in agricultural soil, ISME J., 5, 1771–1783, 2011.
Griffiths, B. S. and Philippot, L.: Insights into the resistance and resilience of the soil microbial community, Fems Microbiol. Rev., 37, 112–129, 2013.
Guillaume, T., Maranguit, D., Murtilaksono, K., and Kuzyakov, Y.: Sensitivity and resistance of soil fertility indicators to land-use changes: New concept and examples from conversion of Indonesian rainforest to plantations, Ecol. Indic., 67, 49–57, https://doi.org/10.1016/j.ecolind.2016.02.039, 2016.
Hannula, S. E. and van Veen, J. A.: Primer sets developed for functional genes reveal shifts in functionality of fungal community in soils, Front. Microbiol., 7, 1897, https://doi.org/10.3389/fmicb.2016.01897, 2016.
Hartmann, A., Schmid, M., van Tuinen, D., and Berg, G.: Plant-driven selection of microbes, Plant Soil, 321, 235–257, 2009.
Hayat, R., Ali, S., Amara, U., Khalid, R., and Ahmed, I.: Soil beneficial bacteria and their role in plant growth promotion: A review, Ann. Microbiol., 60, 579–598, https://doi.org/10.1007/s13213-010-0117-1, 2010.
Haynes, R. J.: Nature of the belowground ecosystem and its development during pedogenesis, Adv. Agron., 127, 43–109, 2014.
Henry, S., Baudoin, E., Lopez-Gutiérrez, J.-C., Martin-Laurent, F., Brauman, A., and Philippot, L.: Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR, J. Microbiol. Meth., 59, 327–335, 2004.
Herrmann, A. M., Coucheney, E., and Nunan, N.: Isothermal microcalorimetry provides new insight into terrestrial carbon cycling, Environ. Sci. Technol., 48, 4344–4352, https://doi.org/10.1021/es403941h, 2014.
Hirsch, P. R., Mauchline, T. H., and Clark, I. M.: Culture-independent molecular techniques for soil microbial ecology, Soil Biol. Biochem., 42, 878–887, https://doi.org/10.1016/j.soilbio.2010.02.019, 2010.
ISO 11063: Soil quality – Method to directly extract DNA from soil samples, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/50025.html (last access: 27 January 2020), 2012.
ISO 11266: Soil quality – Guidance on laboratory testing for biodegradation of organic chemicals in soil under aerobic conditions, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/19244.html (last access: 27 January 2020), 1994.
ISO 14238: Soil quality – Biological methods – Determination of nitrogen mineralization and nitrification in soils and the influence of chemicals on these processes, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/56033.html (last access: 27 January 2020), 2012.
ISO 14239: Soil quality – Laboratory incubation systems for measuring the mineralization of organic chemicals in soil under aerobic conditions, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/69583.html (last access: 27 January 2020), 2017.
ISO 14240-1: Soil quality – Determination of soil microbial biomass – Part 1: Substrate-induced respiration method, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/21530.html (last access: 27 January 2020), 1997.
ISO 14240-2: Soil quality – Determination of soil microbial biomass – Part 2: Fumigation-extraction method, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/23951.html (last access: 27 January 2020), 1997.
ISO 15473: Soil quality – Guidance on laboratory testing for biodegradation of organic chemicals in soil under anaerobic conditions, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/27189.html (last access: 27 January 2020), 2002.
ISO 15685: Soil quality – Determination of potential nitrification and inhibition of nitrification – Rapid test by ammonium oxidation, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/53530.html (last access: 27 January 2020), 2012.
ISO 16072: Soil quality – Laboratory methods for determination of microbial soil respiration, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/32096.html (last access: 27 January 2020), 2002.
ISO 17155: Soil quality – Determination of abundance and activity of soil microflora using respiration curves, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/53529.html (last access: 27 January 2020), 2012.
ISO 17601: Soil quality – Estimation of abundance of selected microbial gene sequences by quantitative PCR from DNA directly extracted from soil, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/60106.html (last access: 27 January 2020), 2016.
ISO 18311: Soil quality – Method for testing effects of soil contaminants on the feeding activity of soil dwelling organisms – Bait-lamina test, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/62102.html (last access: 27 January 2020), 2016.
ISO 23753-1: Soil quality – Determination of dehydrogenase activity in soils – Part 1: Method using triphenyltetrazolium chloride (TTC), International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/70145.html (last access: 27 January 2020), 2019.
ISO 23753-2: Soil quality – Determination of dehydrogenase activity in soils – Part 2: Method using iodotetrazolium chloride (INT), International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/70146.html (last access: 27 January 2020), 2019.
ISO 20130: Soil quality – Measurement of enzyme activity patterns in soil samples using colorimetric substrates in micro-well plates, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/67074.html (last access: 27 January 2020), 2018.
ISO 20951: Soil Quality – Guidance on methods for measuring greenhouse gases (CO2, N2O, CH4) and ammonia (NH3) fluxes between soils and the atmosphere, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/69534.html (last access: 27 January 2020), 2018.
ISO/CD 23265: Soil quality – Test for measuring organic matter decomposition in contaminated soil, International Organization for Standardization, Geneva, Switzerland, under development, available at: https://www.iso.org/standard/75115.html (last access: 27 January 2020), 2018.
ISO/TS 20131-1: Soil quality – Easy laboratory assessments of soil denitrification, a process source of N2O emissions – Part 1: Soil denitrifying enzymes activities, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/67075.html (last access: 27 January 2020), 2018.
ISO/TS 20131-2: Soil quality – Easy laboratory assessments of soil denitrification, a process source of N2O emissions – Part 2: Assessment of the capacity of soils to reduce N2O, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/67076.html (last access: 27 January 2020), 2018.
ISO/TS 22939: Soil quality – Measurement of enzyme activity patterns in soil samples using fluorogenic substrates in micro-well plates, International Organization for Standardization, Geneva, Switzerland, available at: https://www.iso.org/standard/41226.html (last access: 27 January 2020), 2019.
ISO/TS 29843-1: Soil quality – Determination of soil microbial diversity – Part 1: Method by phospholipid fatty acid analysis (PLFA) and phospholipid ether lipids (PLEL) analysis, International Organization for Standardization, Geneva, available at: https://www.iso.org/standard/45703.html (last access: 27 January 2020), 2010.
ISO/TS 29843-2: Soil quality – Determination of soil microbial diversity – Part 2: Method by phospholipid fatty acid analysis (PLFA) using the simple PLFA extraction method, International Organization for Standardization, Geneva, available at: https://www.iso.org/standard/54070.html (last access: 27 January 2020), 2011.
Jänsch, S., Scheffczyk, A., and Römbke, J.: The bait lamina test – A possible screening method for earthworm toxicity testing, Euro.-Mediterr. J. Environ. Integr., 2, 5, https://doi.org/10.1007/s41207-017-0015-z, 2017.
Janos, D. P., Garamszegi, S., and Beltran, B.: Glomalin extraction and measurement, Soil Biol. Biochem., 40, 728–739, https://doi.org/10.1016/j.soilbio.2007.10.007, 2008.
Jansson, J. K. and Hofmockel, K. S.: The soil microbiome – from metagenomics to metaphenomics, Curr. Opin. Microbiol., 43, 162–168, https://doi.org/10.1016/j.mib.2018.01.013, 2018.
Jeffries, T. C., Rayu, S., Nielsen, U. N., Lai, K., Ijaz, A., Nazaries, L., and Singh, B. K.: Metagenomic functional potential predicts degradation rates of a model organophosphorus xenobiotic in pesticide contaminated soils, Front. Microbiol., 9, 147, https://doi.org/10.3389/fmicb.2018.00147, 2018.
Jiang, X., Cao, L., Zhang, R., Yan, L., Mao, Y., and Yang, Y.: Effects of nitrogen addition and litter properties on litter decomposition and enzyme activities of individual fungi, Appl. Soil Ecol., 80, 108–115, 2014.
Joergensen, R. G. and Emmerling, C.: Methods for evaluating human impact on soil microorganisms based on their activity, biomass, and diversity in agricultural soils, J. Plant Nutr. Soil Sc., 169, 295–309, https://doi.org/10.1002/jpln.200521941, 2006.
Jung, J., Choi, S., Jung, H., Scow, K. M., and Park, W.: Primers for amplification of nitrous oxide reductase genes associated with Firmicutes and Bacteroidetes in organic-compound-rich soils, Microbiology, 159, 307–315, https://doi.org/10.1099/mic.0.060194-0, 2013.
Keuskamp, J. A., Dingemans, B. J. J., Lehtinen, L., Sarneel, J. M., and Hefting, M. M.: Tea Bag Index: a novel approach to collect uniform decomposition data across ecosystems, Methods Ecol. Evol., 4, 1070–1075, 2013.
Knacker, T., Förster, B., Römbke, J., and Frampton, G. K.: Assessing the effects of plant protection products on organic matter breakdown in arable fields – Litter decomposition test systems, Soil Biol. Biochem., 35, 1269–1287, https://doi.org/10.1016/S0038-0717(03)00219-0, 2003.
Kolb, S., Knief, C., Stubner, S., and Conrad, R.: Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays, Appl. Environ. Microb., 69, 2423–2429, https://doi.org/10.1128/AEM.69.5.2423-2429.2003, 2003.
Kuffner, M., Hai, B., Rattei, T., Melodelima, C., Schloter, M., Zechmeister-Boltenstern, S., Jandl, R., Schindlbacher, A., and Sessitsch, A.: Effects of season and experimental warming on the bacterial community in a temperate mountain forest soil assessed by 16S rRNA gene pyrosequencing, FEMS Microbiol. Ecol., 82, 551–562, https://doi.org/10.1111/j.1574-6941.2012.01420.x, 2012.
Kvas, S., Rahn, J., Engel, K., Neufeld, J. D., Villeneuve, P. J., Trevors, J. T., Lee, H., Scroggins, R. P., and Beaudette, L. A.: Development of a microbial test suite and data integration method for assessing microbial health of contaminated soil, J. Microbiol. Meth., 143, 66–77, 2017.
Levy-Booth, D. J., Prescott, C. E., and Grayston, S. J.: Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems, Soil Biol. Biochem., 75, 11–25, https://doi.org/10.1016/j.soilbio.2014.03.021, 2014.
Liu, W., Qiao, C., Yang, S., Bai, W., and Liu, L.: Microbial carbon use efficiency and priming effect regulate soil carbon storage under nitrogen deposition by slowing soil organic matter decomposition, Geoderma, 332, 37–44, https://doi.org/10.1016/j.geoderma.2018.07.008, 2018.
Lueders, T., Manefield, M., and Friedrich, M. W.: Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients, Environ. Microbiol., 6, 73–78, 2004.
Lugtenberg, B. and Kamilova, F.: Plant-growth-promoting rhizobacteria, Annu. Rev. Microbiol., 63, 541–556, https://doi.org/10.1146/annurev.micro.62.081307.162918, 2009.
Martin-Laurent, F., Cornet, L., Ranjard, L., Lopez-Gutiérrez, J. C., Philippot, L., Schwartz, C., Chaussod, R., Catroux, G., and Soulas, G.: Estimation of atrazine-degrading genetic potential and activity in three French agricultural soils, FEMS Micobiol. Ecol., 48, 425–435, 2004.
MEA (Millennium Ecosystem Assessment): Ecosystems and Human Well-being: Synthesis, Island Press, Washington, D.C., 2005.
Modrzyński, J. J., Christensen, J. H., Mayer, P., and Brandt, K. K.: Limited recovery of soil microbial activity after transient exposure to gasoline vapors, Environ. Pollut., 216, 826–835, https://doi.org/10.1016/j.envpol.2016.06.054, 2016.
Montgomery, D. R.: Dirt: The Erosion of Civilizations, University of California Press, Berkeley, Calif., London, 2008.
Nannipieri, P. and Eldor, P.: The chemical and functional characterization of soil N and its biotic components, Soil Biol. Biochem., 41, 2357–2369, https://doi.org/10.1016/j.soilbio.2009.07.013, 2009.
Nannipieri, P., Greco, S., and Ceccanti, B.: Ecological Significance of the Biological Activity in Soil, in: Soil Biochemistry, Volume 6, edited by: Bollag, J. M. and Stotzky, G., Routledge, New York, 293–355, 2017.
Nicol, G. W., Glover, L. A., and Prosser, J. I.: Spatial analysis of archaeal community structure in grassland soil, Appl. Environ. Microb., 69, 7420–7429, https://doi.org/10.1128/AEM.69.12.7420-7429.2003, 2003.
Nienstedt, K., Brock, T. C. M., Van Wensem, J., Montforts, M., Hart, A., Aagaard, A., Alix, A., Boesten, J., Bopp, S. K., Brown, C., Capri, E., Forbes, V., Köpp, H., Liess, M., Luttik, R., Maltby, L., Sousa, J.-P., Streissl, F., and Hardy, A. R.: Development of a framework based on an ecosystem services approach for deriving specific protection goals for environmental risk assessment of pesticides, Sci. Total Environ., 415, 31–38, 2012.
Norton, J. M., Alzerreca, J. J., Suwa, Y., and Klotz, M. G.: Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria, Arch. Microbiol., 177, 139–149, https://doi.org/10.1007/s00203-001-0369-z, 2002.
Ockleford, C., Adriaanse, P., Berny, P., Brock, T., Duquesne, S., Grilli, S., Hernandez-Jerez, A. F., Bennekou, S. H., Klein, M., Kuhl, T., Laskowski, R., Machera, K., Pelkonen, O., Pieper, S., Stemmer, M., Sundh, I., Teodorovic, I., Tiktak, A., Topping, C. J., Wolterink, G., Craig, P., de Jong, F., Manachini, B., Sousa, P., Swarowsky, K., Auteri, D., Arena, M., and Rob, S.: EFSA PPR Panel (EFSA Panel on Plant Protection Products and their Residues) Scientific Opinion addressing the state of the science on risk assessment of plant protection products for in-soil organisms, EFSA Journal, 15, 255, https://doi.org/10.2903/j.efsa.2017.4690, 2017.
OECD: Test No. 216: Soil Microorganisms: Nitrogen Transformation Test, Paris, France, 2000a.
OECD: Test No. 217: Soil Microorganisms: Carbon Transformation Test, Paris, France, 2000b.
OECD: OECD Series on Testing and Assessment No. 56, Guidance document on the breakdown of organic matter in litterbags, ENV/JM/MONO 23, Paris, France, 2006.
Ollivier, J., Kleineidam, K., Reichel, R., Thiele-Bruhn, S., Kotzerke, A., Kindler, R., Wilke, B. M., and Schloter, M.: Effect of sulfadiazine-contaminated pig manure on the abundances of genes and transcripts involved in nitrogen transformation in the root-rhizosphere complexes of maize and clover, Appl. Environ. Microb., 76, 7903–7909, 2010.
Ouyang, Y., Reeve, J. R., and Norton, J. M.: Soil enzyme activities and abundance of microbial functional genes involved in nitrogen transformations in an organic farming system, Biol. Fert. Soils, 54, 437–450, 2018.
Pankhurst, C. E., Yu, S., Hawke, B. G., and Harch, B. D.: Capacity of fatty acid profiles and substrate utilization patterns to describe differences in soil microbial communities associated with increased salinity or alkalinity at three locations in South Australia, Biol. Fert. Soils, 33, 204–217, 2001.
Parker, S. S.: Buried treasure: Soil biodiversity and conservation, Biodivers. Conserv., 19, 3743–3756, 2010.
Paul, E. A.: Soil Microbiology, Ecology and Biochemistry, 4th Edn., Academic Press, Burlington, MA, 2015.
Peng, J., Lü, Z., Rui, J., and Lu, Y.: Dynamics of the methanogenic archaeal community during plant residue decomposition in an anoxic rice field soil, Appl. Environ. Microb., 74, 2894–2901, https://doi.org/10.1128/AEM.00070-08, 2008.
Penton, C. R., Gupta, V. V. S. R., Yu, J., and Tiedje, J. M.: Size matters: Assessing optimum soil sample size for fungal and bacterial community structure analyses using high throughput sequencing of rRNA gene amplicons, Front. Microbiol., 7, 824, https://doi.org/10.3389/fmicb.2016.00824, 2016.
Philippot, L., Ritz, K., Pandard, P., Hallin, S., and Martin-Laurent, F.: Standardisation of methods in soil microbiology: Progress and challenges, FEMS Microbiol. Ecol., 82, 1–10, https://doi.org/10.1111/j.1574-6941.2012.01436.x, 2012.
Prado, A. G. S. and Airoldi, C.: The effect of the herbicide diuron on soil microbial activity, Pest Manag. Sci., 57, 640–644 https://doi.org/10.1002/ps.321, 2001.
Prado, A. G. S. and Airoldi, C.: A toxicity view of the pesticide picloram when immobilized onto a silica gel surface, Anal. Bioanal. Chem., 376, 686–690, https://doi.org/10.1007/s00216-003-1927-9, 2003.
Preston-Mafham, J., Boddy, L., and Randerson, P. F.: Analysis of microbial community functional diversity using sole-carbon-source utilisation profiles – A critique, FEMS Microbiol. Ecol., 42, 1–14, https://doi.org/10.1016/S0168-6496(02)00324-0, 2002.
Pulleman, M., Creamer, R., Hamer, U., Helder, J., Pelose, C., Peres, G., and Rutgers, M.: Soil biodiversity, biological indicators and soil ecosystem services – an overview of European approaches, Curr. Opin. Env. Sust., 4, 529–538, 2012.
Quince, C., Walker, A. W., Simpson, J. T., Loman, N. J., and Segata, N.: Shotgun metagenomics, from sampling to analysis, Nat. Biotechnol., 35, 833–844, 2017.
Ramirez, K. S., Knight, C. G., De Hollander, M., Brearley, F. Q., Constantinides, B., Cotton, A., Creer, S., Crowther, T. W., Davison, J., Delgado-Baquerizo, M., Dorrepaal, E., Elliott, D. R., Fox, G., Griffiths, R. I., Hale, C., Hartman, K., Houlden, A., Jones, D. L., Krab, E. J., Maestre, F. T., McGuire, K. L., Monteux, S., Orr, C. H., Van Der Putten, W. H., Roberts, I. S., Robinson, D. A., Rocca, J. D., Rowntree, J., Schlaeppi, K., Shepherd, M., Singh, B. K., Straathof, A. L., Bhatnagar, J. M., Thion, C., Van Der Heijden, M. G. A., and De Vries, F. T.: Detecting macroecological patterns in bacterial communities across independent studies of global soils, Nat. Microbiol., 3, 189–196, 2018.
Regan, K. M., Nunan, N., Boeddinghaus, R. S., Baumgartner, V., Berner, D., Boch, S., Oelmann, Y., Overmann, J., Prati, D., Schloter, M., Schmitt, B., Sorkau, E., Steffens, M., Kandeler, E., and Marhan, S.: Seasonal controls on grassland microbial biogeography: Are they governed by plants, abiotic properties or both?, Soil Biol. Biochem., 71, 21–30, https://doi.org/10.1016/j.soilbio.2013.12.024, 2014.
Ribbons, R. R., Levy-Booth, D. J., Masse, J., Grayston, S. J., McDonald, M. A., Vesterdal, L., and Prescott, C. E.: Linking microbial communities, functional genes and nitrogen-cycling processes in forest floors under four tree species, Soil Biol. Biochem., 103, 181–191, https://doi.org/10.1016/j.soilbio.2016.07.024, 2016.
Rillig, M. C.: Arbuscular mycorrhizae, glomalin, and soil aggregation, Can. J. Soil Sci., 84, 355–363, https://doi.org/10.4141/S04-003, 2004.
Rillig, M. C. and Mummey, D. L.: Mycorrhizas and soil structure, New Phytol., 171, 41–53, https://doi.org/10.1111/j.1469-8137.2006.01750.x, 2006.
Ritz, K., Black, H. I. J., Campbell, C. D., Harris, J. A., and Wood, C.: Selecting biological indicators for monitoring soils: A framework for balancing scientific and technical opinion to assist policy development, Ecol. Indic., 9, 1212–1221, 2009.
Römbke, J., Jänsch, S., Meier, M., Hilbeck, A., Teichmann, H., and Tappeser, B.: General recommendations for soil ecotoxicological tests suitable for the environmental risk assessment (ERA) of genetically modified plants (GMPs), Integr. Environ. Assess., 6, 287–300, 2010.
Rousk, J.: Biomass or growth? How to measure soil food webs to understand structure and function, Soil Biol. Biochem., 102, 45–47, https://doi.org/10.1016/j.soilbio.2016.07.001, 2016.
Rousk, J., Demoling, L. A., and Bååth, E.: Contrasting short-term antibiotic effects on respiration and bacterial growth compromises the validity of the selective respiratory inhibition technique to distinguish fungi and bacteria, Microb. Ecol., 58, 75–85, https://doi.org/10.1007/s00248-008-9444-1, 2009a.
Rousk, J., Brookes, P. C., and Bååth, E.: Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization, Appl. Environ. Microb., 75, 1589–1596, https://doi.org/10.1128/AEM.02775-08, 2009b.
Rutgers, M., Wouterse, M., Drost, S. M., Breure, A. M., Mulder, C., Stone, D., Creamer, R. E., Winding, A., and Bloem, J.: Monitoring soil bacteria with community-level physiological profiles using Biolog™ ECO-plates in the Netherlands and Europe, Appl. Soil Ecol., 97, 23–35, https://doi.org/10.1016/j.apsoil.2015.06.007, 2016.
Sakakibara, S. M., Jones, M. D., Gillespie, M., Hagerman, S. M., Forrest, M. E., Simard, S. W., and Durall, D. M.: A comparison of ectomycorrhiza identification based on morphotyping and PCR-RFLP analysis, Mycol. Res., 106, 868–878, 2002.
Savazzini, F., Longa, C. M., Pertot, I., and Gessler, C. J.: Real-time PCR for detection and quantification of the biocontrol agent Trichoderma atroviride strain SC1 in soil, Microbiological Methods, 73, 185–194, https://doi.org/10.1016/j.mimet.2008.02.004, 2008.
Scheu, S., Schlitt, N., Tiunov, A. V., Newington, J. E., and Jones, T. H.: Effects of the presence and community composition of earthworms on microbial community functioning, Oecologia, 133, 254–260, https://doi.org/10.1007/s00442-002-1023-4, 2002.
Scheu, S., Ruess, L., and Bonkowski, M.: Interactions between microorganisms and soil micro- and mesofauna, in: Microorganisms in Soils: Roles in Genesis and Functions, edited by: Varma, A. and Buscot, F., Vol. 3, Springer, Berlin, Heidelberg, 253–275, 2005.
Schmitt, H., Van Beelen, P., Tolls, J., and Van Leeuwen, C. L.: Pollution-induced community tolerance of soil microbial communities caused by the antibiotic sulfachloropyridazine, Environ. Sci. Technol., 38, 1148–1153, 2004.
Schöler, A., Jacquiod, S., Vestergaard, G., Schulz, S., and Schloter, M.: Analysis of soil microbial communities based on amplicon sequencing of marker genes, Biol. Fert. Soils, 53, 485–489, 2017.
Schwartz, E.: Characterization of growing microorganisms in soil by stable isotope probing with , Appl. Environ. Microb., 73, 2541–2546, 2007.
Schwartz, E., Hayer, M., Hungate, B. A., Koch, B. J., McHugh, T. A., Mercurio, W., Morrissey, E. M., and Soldanova, K.: Stable isotope probing with 18O-water to investigate microbial growth and death in environmental samples, Curr. Opin. Biotech., 41, 14–18, 2016.
Schulz, S., Bergkemper, F., De Vries, M., Schöler, A., and Schloter, M.: qPCR for quantitative validation of metagenomic data [qPCR zur quantitativen Validierung von Metagenomdaten], BioSpektrum, 22, 265–269, https://doi.org/10.1007/s12268-016-0684-1, 2016.
Sessitsch, A., Hackl, E., Wenzl, P., Kilian, A., Kostic, T., Stralis-Pavese, N., Sandjong, B. T., and Bodrossy, L.: Diagnostic microbial microarrays in soil ecology, New Phytol., 171, 719–736, https://doi.org/10.1111/j.1469-8137.2006.01824.x, 2006.
Singh, B. K., Tate, K. R., Kolipaka, G., Hedley, C. B., Macdonald, C. A., Millard, P., and Murrell, J. C.: Effect of afforestation and reforestation of pastures on the activity and population dynamics of methanotrophic bacteria, Appl. Environ. Microb., 73, 5153–5161, https://doi.org/10.1128/AEM.00620-07, 2007.
Six, J., Bossuyt, H., Degryze, S., and Denef, K.: A history of research on the link between (micro)aggregates, soil biota, and soil organic matter dynamics, Soil Till. Res., 79, 7–31 https://doi.org/10.1016/j.still.2004.03.008, 2004.
Smith, C. J. and Osborn, A. M.: Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology, FEMS Microbiol. Ecol., 67, 6–20, https://doi.org/10.1111/j.1574-6941.2008.00629.x, 2009.
Šnajdr, J., Valášková, V., Merhautová, V., Herinková, J., Cajthaml, T., and Baldrian, P.: Spatial variability of enzyme activities and microbial biomass in the upper layers of Quercus petraea forest soil, Soil Biol. Biochem., 40, 2068–2075, 2008.
Spohn, M., Pötsch, E. M., Eichorst, S. A., Woebken, D., Wanek, W., and Richter, A.: Soil microbial carbon use efficiency and biomass turnover in a long-term fertilization experiment in a temperate grassland, Soil Biol. Biochem., 97, 168–175, https://doi.org/10.1016/j.soilbio.2016.03.008, 2016.
Steinberg, L. M. and Regan, J. M.: mcrA-targeted real-time quantitative PCR method to examine methanogen communities, Appl. Environ. Microb., 75, 4435–4442, 2009.
Strickland, M. S. and Rousk, J.: Considering fungal: Bacterial dominance in soils – Methods, controls, and ecosystem implications, Soil Biol. Biochem., 42, 1385–1395, https://doi.org/10.1016/j.soilbio.2010.05.007, 2010.
Suriyavirun, N., Krichels, A. H., Kent, A. D., and Yang, W. H.: Microtopographic differences in soil properties and microbial community composition at the field scale, Soil Biol. Biochem., 131, 71–80, https://doi.org/10.1016/j.soilbio.2018.12.024, 2019.
Takriti, M., Wild, B., Schnecker, J., Mooshammer, M., Knoltsch, A., Lashchinskiy, N., Eloy Alves, R. J., Gentsch, N., Gittel, A., Mikutta, R., Wanek, W., and Richter, A.: Soil organic matter quality exerts a stronger control than stoichiometry on microbial substrate use efficiency along a latitudinal transect, Soil Biol. Biochem., 121, 212–220, https://doi.org/10.1016/j.soilbio.2018.02.022, 2018.
TEEB: The Economics of Ecosystems and Biodiversity Ecological and Economic Foundations, edited by: Kumar, P., Earthscan, Routledge, London, UK, 2010.
Thiele-Bruhn, S., Bloem, J., de Vries, F. T., Kalbitz, K., and Wagg, C.: Linking soil biodiversity and agricultural soil management, Curr. Opin. Env. Sust., 4, 523–528, 2012.
Tian, J., He, N., Hale, L., Niu, S., Yu, G., Liu, Y., Blagodatskaya, E., Kuzyakov, Y., Gao, Q., and Zhou, J.: Soil organic matter availability and climate drive latitudinal patterns in bacterial diversity from tropical to cold temperate forests, Funct. Ecol., 32, 61–70, https://doi.org/10.1111/1365-2435.12952, 2018.
Traugott, M., Kamenova, S., Ruess, L., Seeber, J., and Plantegenest, M.: Empirically characterising trophic networks: What emerging DNA-based methods, stable isotope and fatty acid analyses can offer, Adv. Ecol. Res., 49, 177–224, 2013.
Van der Putten, W. H., Mudgal, S., Turbé, A., Toni, A. D., Lavelle, P., Benito, P., and Ruiz, N.: Soil biodiversity: functions, threats and tools for policy makers, Bio Intelligence Service, Paris, France, 2010.
Voříšková, A., Jansa, J., Püschel, D., Krüger, M., Cajthaml, T., Vosátka, M., and Janoušková, M.: Real-time PCR quantification of arbuscular mycorrhizal fungi: does the use of nuclear or mitochondrial markers make a difference?, Mycorrhiza, 27, 577–585, https://doi.org/10.1007/s00572-017-0777-9, 2017.
Wagg, C., Bender, S. F., Widmer, F., and Van Der Heijden, M. G. A.: Soil biodiversity and soil community composition determine ecosystem multifunctionality, P. Natl. Acad. Sci. USA, 111, 5266–5270, https://doi.org/10.1073/pnas.1320054111, 2014.
Wallenstein, M. D., Myrold, D. D., Firestone, M., and Voytek, M.: Environmental controls on denitrifying communities and denitrification rates: Insights from molecular methods, Ecol. Appl., 16, 2143–2152, https://doi.org/10.1890/1051-0761(2006)016[2143:ECODCA]2.0.CO;2, 2006.
Wardle, D. A. and Ghani, A.: A critique of the microbial metabolic quotient (qCO2) as a bioindicator of disturbance and ecosystem development, Soil Biol. Biochem., 27, 1601–1610, https://doi.org/10.1016/0038-0717(95)00093-T, 1995.
Winding, A. and Hendriksen, N. B.: Comparison of CLPP and enzyme activity assay for functional characterization of bacterial soil communities, J. Soil. Sediment., 7, 411–417, https://doi.org/10.1065/jss2007.11.262, 2007.
Wright, S. F., Upadhyaya, A., and Buyer, J. S.: Comparison of N-linked oligosaccharides of glomalin from arbuscular mycorrhizal fungi and soils by capillary electrophoresis, Soil Biol. Biochem., 30, 1853–1857, https://doi.org/10.1016/S0038-0717(98)00047-9, 1998.
Xia, W., Zhang, C., Zeng, X., Feng, Y., Weng, J., Lin, X., Zhu, J., Xiong, Z., Xu, J., Cai, Z., and Jia, Z.: Autotrophic growth of nitrifying community in an agricultural soil, ISME J., 5, 1226–1236, https://doi.org/10.1038/ismej.2011.5, 2011.
Xu, S., Feng, S., Sun, H., Wu, S., Zhuang, G., Deng, Y., Bai, Z., Jing, C., and Zhuang, X.: Linking N2O emissions from biofertilizer-amended soil of tea plantations to the abundance and structure of N2O-reducing microbial communities, Environ. Sci. Technol., 52, 11338–11345, https://doi.org/10.1021/acs.est.8b04935, 2018.
Xue, K., Zhou, J., Van Nostrand, J., Mench, M., Bes, C., Giagnoni, L., and Renella, G.: Functional activity and functional gene diversity of a Cu-contaminated soil remediated by aided phytostabilization using compost, dolomitic limestone and a mixed tree stand, Environ. Pollut., 242, 229–238, 2018.
Yates, M. V., Nakatsu, C. H., Miller, R. V., and Pillai, S. D.: Manual of Environmental Microbiology, 4th Edn., American Society of Microbiology, Washington DC, USA, 2016.
Yergeau, E., Hogues, H., Whyte, L. G., and Greer, C. W.: The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR and microarray analyses, ISME J., 4, 1206–1214, https://doi.org/10.1038/ismej.2010.41, 2010.
Zhang, K., Li, X., Cheng, X., Zhang, Z., and Zhang, Q.: Changes in soil properties rather than functional gene abundance control carbon and nitrogen mineralization rates during long-term natural revegetation, Plant Soil, 443, 293–306, 2019.





