Abstract
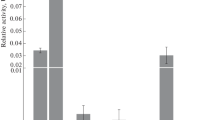
Nonrandom distribution of meiotic recombination events along the chromosomes shapes the diversity of potential genetic combinations among the offspring. To redistribute the chromosomal regions involved in recombination events, it was proposed to use meiosis-specific genes of Spo11 proteins (generating double-strand DNA breaks) from phylogenetically different organisms. For these purposes, transgenic tomato plants expressing native SPO11 genes from yeast (Saccharomyces cerevisae) or Arabidopsis thaliana under the control of constitutive 35S CaMV promoter were constructed. Genetic analysis showed that expression of both target SPO11 genes partly disturbed the monogenic inheritance of marker Wv:wv alleles in chromosome 2 and suppressed the crossing over in the region between the wv and d genes. A stable negative correlation between the target gene expression levels and the decrease in the frequency of crossing over in the analyzed chromosomal region was found. The possible genetic mechanisms underlying the redistribution of crossovers along chromosome 2 resulting from the target SPO11 gene expression are discussed.




Similar content being viewed by others
REFERENCES
Keeney, S., Spo11 and the formation of DNA double-strand breaks in meiosis, Genome Dyn. Stab., 2008, vol. 2, pp. 81—123. https://doi.org/10.1007/7050_2007_026
Vrielynck, N., Chambon, A., Vezon, D., et al., A DNA topoisomerase VI–like complex initiates meiotic recombination, Science, 2016, vol. 351, pp. 939—943. https://doi.org/10.1126/science.aad5196
Robert, T., Nore, A., Brun, C., et al., The TopoVIB-like protein family is required for meiotic DNA double-strand break formation, Science, 2016, vol. 351, pp. 943—949. https://doi.org/10.1126/science.aad5309
De Boer, E., Jasin, M., and Keeney, S., Analysis of recombinants in female mouse meiosis, Methods Mol. Biol., 2013, vol. 957, pp. 19—45. https://doi.org/10.1007/978-1-62703-191-2_2
Choi, K., Zhao, X., Kelly, K.A., et al., Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters, Nat. Genet., 2013, vol. 45, pp. 1327—1336. https://doi.org/10.1038/ng.2766
Lam, I. and Keeney, S., Mechanism and regulation of meiotic recombination initiation, Cold Spring Harbor Perspect. Biol., 2014, vol. 7, no. 1. a016634. https://doi.org/10.1101/cshperspect.a016634
Goldfarb, T. and Lichten, M., Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis, PLoS Biol., 2010, vol. 8, no. 10. e1000520. https://doi.org/10.1371/journal.pbio.1000520
Cole, F., Keeney, S., and Jasin, M., Preaching about the converted: how meiotic gene conversion influences genomic diversity, Ann. N.Y. Acad. Sci., 2012, vol. 1267, pp. 95—102. https://doi.org/10.1111/j.1749-6632.2012.06595.x
Crismani, W., Girard, C., Froger, N., et al., FANCM limits meiotic crossovers, Science, 2012, vol. 336, pp. 1588—1590. https://doi.org/10.1126/science.1220381
Girard, C., Chelysheva, L., Choinard, S., et al., AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms, PLoS Genet., 2015, vol. 11, no. 7. e1005369. https://doi.org/10.1371/journal.pgen.1005369
Seguela-Arnaud, M., Crismani, W., Larcheveque, C., et al., Multiple mechanisms limit meiotic crossovers: TOP3alpha and two BLM homologs antagonize crossovers in parallel to FANCM, Proc. Natl. Acad. Sci. U.S.A., 2015, vol. 112, pp. 4713—4718. https://doi.org/10.1073/pnas.1423107112
Ziolkowski, P.A., Underwood, C.J., Lambing, C., et al., Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination, Genes Dev., 2017, vol. 31, pp. 306—317. https://doi.org/10.1101/gad.295501.116
Komakhin, R.A., Komakhina, V.V., Milyukova, N.A. et al., Analysis of the meiotic recombination frequency in transgenic tomato hybrids expressing recA and NLS-recA-licBM3 genes, Russ. J. Genet., 2012, vol. 48, no. 1, pp. 23—31. https://doi.org/10.1134/S1022795411110093
Peciña, A., Smith, K., Mézard, C., et al., Targeted stimulation of meiotic recombination, Cell, 2002, vol. 111, no. 2, pp. 173—184. https://doi.org/10.1016/S0092-8674(02)01002-4
Sarno, R., Vicq, Y., Uematsu, N., et al., Programming sites of meiotic crossovers using Spo11 fusion proteins, Nucleic Acids Res., 2017, vol. 45, no. 19. e164. https://doi.org/10.1093/nar/gkx739
Grelon, M., Vezon, D., Gendrot, G., and Pelletier, G., AtSPO11-1 is necessary for efficient meiotic recombination in plants, EMBO J., 2001, vol. 20, pp. 589—600. https://doi.org/10.1093/emboj/20.3.589
Baudat, F., Manova, K., Yuen, J.P., et al., Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11, Mol. Cell, 2000, vol. 6, pp. 989—998. https://doi.org/10.1016/S1097-2765(00)00098-8
Kauppi, L., Barchi, M., Lange, J., et al., Numerical constraints and feedback control of double-strand breaks in mouse meiosis, Genes Dev., 2013, vol. 27, pp. 873—886. https://doi.org/10.1101/gad.213652.113
Komakhin, R.A., Komakhina, V.V. Compartmentalization of Spo11p in vegetative cells of yeast Saccharomyces cerevisiae, Mol. Biol. (Moscow), 2008, vol. 42, no. 3, pp. 436—441. https://doi.org/10.1134/S0026893308030126
Komakhin, R.A., Komakhina, V.V., Milyukova, N.A., et al., Transgenic tomato plants expressing resA and NLS-resA-lisVM3 genes as a model for studying meiotic recombination, Russ. J. Genet., 2010, vol. 46, no. 12, pp. 1440—1448. https://doi.org/10.1134/S1022795410120069
Komakhin, R.A., Strelnikova, S.R., and Zhuchenko, A.A., Genetic characteristics of the marker line for cultivated tomato Mo938, Russ. J. Genet., 2019, vol. 55, no. 1, pp. 60—69. https://doi.org/10.1134/S1022795419010083
Muller, P.Y., Janovjak, H., Miserez, A.R., and Dobbie, Z., Processing of gene expression data generated by quantitative real-time RT-PCR, Biotechniques, 2002, vol. 32, pp. 1372—1374, 1376, 1378—1379.
Pogorelko, G.V. and Fursova, O.V., A highly efficient miPCR mehtod for isolating FSTs from transgenic Arabidopsis thaliana plants, J. Genet., 2008, vol. 87, pp. 133—140. https://doi.org/10.1007/s12041-008-0020-8
Permyakova, N.V. and Deineko, E.V., Vector DNA fragments integrating into the genome of transgenic carrot plants during agrobacterial transformation, Vestn. Tomsk. Gos. Univ., Biol., 2015, vol. 32, pp. 145—161.
Orlova, N.N., Geneticheskii analiz (Genetic Analysis), Moscow: Moscow Gos. Univ., 1991.
Komakhin, R.A., Milyukova, N.A., Strelnikova, S.R., et al., Inheritance of marker genes among progeny of interspecific tomato hybrids expressing the recA Escherichia coli gene, Russ. J. Genet., 2019, vol. 55, no. 4, pp. 433—443. https://doi.org/10.1134/S1022795419040069
Hammarlund, M., Davis, M.W., Nguyen, H., et al., Heterozygous insertions alter crossover distribution but allow crossover interference in Caenorhabditis elegans, Genetics, 2005, vol. 171, pp. 1047—1056. https://doi.org/10.1534/genetics.105.044834
Yunusov, Z.R., Solov’ev, A.A., Mikhailenko, S.N., et al., Effect of transgenes on meiotic recombination in higher eukaryotes exemplified by tomato, S.-kh. Biol., 2009, vol. 44, pp. 52—59.
Yoshinori, S., Tokai, T., Agawa, Y., et al., The double-stranded break-forming activity of plant SPO11s and a novel rice SPO11 revealed by a Drosophila bioassay, BMC Mol. Biol., 2012, vol. 13, no. 1. https://doi.org/10.1186/1471-2199-13-1
Murakami, H. and Nicolas, A., Locally, meiotic double-strand breaks targeted by Gal4BD-Spo11 occurs at discrete sites with a sequence preference, Mol. Cell Biol., 2009, vol. 29, pp. 3500—3516. https://doi.org/10.1128/MCB.00088-09
Jeffreys, A. and Neumann, R., Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot, Nat. Genet., 2002, vol. 31, pp. 267—271. https://doi.org/10.1038/ng910
Anderson, L.K., Lohmiller, L.D., Tang, X., et al., Combined fluorescent and electron microscopic imaging unveils the specific properties of two classes of meiotic crossovers, Proc. Natl. Acad. Sci. U.S.A., 2014, vol. 111, pp. 13415—13420.https://doi.org/10.1073/pnas.1406846111
Funding
Construction of transgenic plants was supported by the Russian Foundation for Basic Research (grant no. 11-04-00873-a); determination of the T-DNA integration locus was carried within the framework of the state contract no. 0574-2019-0001 (state registration no. AAAA-A18-118051890110-1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by N. Maleeva
Rights and permissions
About this article
Cite this article
Komakhina, V.V., Krinitsina, A.A., Milyukova, N.A. et al. Expression of Recombinant SPO11 Genes Locally Alters Crossing Over in Tomato. Russ J Genet 56, 1079–1089 (2020). https://doi.org/10.1134/S1022795420090124
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795420090124