Single-Cell Elasticity Measurement with an Optically Actuated Microrobot
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microtool Design and Fabrication
2.2. Cell Culturing
2.3. HOT Setup
2.4. Cell Indentation Experiment
2.5. Data Analysis
2.6. Trap Stiffness Calibration
3. Results
3.1. Trap Stiffness Calibration
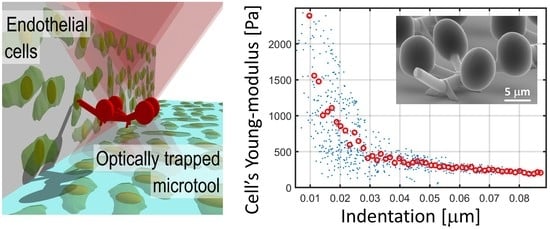
3.2. Endothelial Cells Young’s Modulus
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Chen, H.; Chandrasekar, S.; Sheetz, M.P.; Stossel, T.P.; Nakamura, F.; Yan, J. Mechanical perturbation of filamin A immunoglobulin repeats 20-21 reveals potential non-equilibrium mechanochemical partner binding function. Sci. Rep. 2013, 3, 1642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oroszi, L.; Galajda, P.; Kirei, H.; Bottka, S.; Ormos, P. Direct measurement of torque in an optical trap and its application to double-strand DNA. Phys. Rev. Lett. 2006, 97, 058301. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Pollard, M.R.; Hughes, C.D.; Ward, A.D.; Van Houten, B.; Towrie, M.; Botchway, S.W.; Parker, A.W.; Kad, N.M. Directly interrogating single quantum dot labelled UvrA2 molecules on DNA tightropes using an optically trapped nanoprobe. Sci. Rep. 2015, 5, 18486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aekbote, B.L.; Fekete, T.; Jacak, J.; Vizsnyiczai, G.; Ormos, P.; Kelemen, L. Surface-modified complex SU-8 microstructures for indirect optical manipulation of single cells. Biomed. Opt. Express 2016, 7, 45–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.; Xie, H.; Wei, T.; Chen, S.; Sun, D. Automated Indirect Transportation of Biological Cells with Optical Tweezers and a 3D Printed Microtool. Appl. Sci. 2019, 9, 2883. [Google Scholar] [CrossRef] [Green Version]
- Maruo, S.; Takaura, A.; Saito, Y. Optically driven micropump with a twin spiral microrotor. Opt. Express 2009, 17, 18525–18532. [Google Scholar] [CrossRef]
- Terray, A.; Oakey, J.; Marr, D.W.M. Microfluidic Control Using Colloidal Devices. Science 2002, 296, 1841–1844. [Google Scholar] [CrossRef]
- Vizsnyiczai, G.; Lestyan, T.; Joniova, J.; Aekbote, B.L.; Strejckova, A.; Ormos, P.; Miskovsky, P.; Kelemen, L.; Bano, G. Optically Trapped Surface-Enhanced Raman Probes Prepared by Silver Photoreduction to 3D Microstructures. Langmuir 2015, 31, 10087–10093. [Google Scholar] [CrossRef] [Green Version]
- Mártonfalvi, Z.; Bianco, P.; Naftz, K.; Ferenczy, G.G.; Kellermayer, M. Force generation by titin folding. Protein Sci. 2017, 26, 1380–1390. [Google Scholar] [CrossRef]
- Barbot, A.; Decanini, D.; Hwang, G. On-chip Microfluidic Multimodal Swimmer toward 3D Navigation. Sci. Rep. 2016, 6, 19041. [Google Scholar] [CrossRef]
- Vizsnyiczai, G.; Aekbote, B.L.; Buzas, A.; Grexa, I.; Ormos, P.; Kelemen, L. High accuracy indirect optical manipulation of live cells with functionalized microtools. In Optical Trapping and Optical Micromanipulation Xiii; Dholakia, K., Spalding, G.C., Eds.; SPIE: Bellingham, WA, USA, 2016; Volume 9922, p. 992216. [Google Scholar]
- Vizsnyiczai, G.; Búzás, A.; Lakshmanrao Aekbote, B.; Fekete, T.; Grexa, I.; Ormos, P.; Kelemen, L. Multiview microscopy of single cells through microstructure-based indirect optical manipulation. Biomed. Opt. Express 2020, 11, 945–962. [Google Scholar] [CrossRef] [PubMed]
- Būtaitė, U.G.; Gibson, G.M.; Ho, Y.-L.D.; Taverne, M.; Taylor, J.M.; Phillips, D.B. Indirect optical trapping using light driven micro-rotors for reconfigurable hydrodynamic manipulation. Nat. Commun. 2019, 10, 1215. [Google Scholar] [CrossRef] [PubMed]
- Turlier, H.; Fedosov, D.A.; Audoly, B.; Auth, T.; Gov, N.S.; Sykes, C.; Joanny, J.F.; Gompper, G.; Betz, T. Equilibrium physics breakdown reveals the active nature of red blood cell flickering. Nat. Phys. 2016, 12, 513. [Google Scholar] [CrossRef] [Green Version]
- Bente, K.; Codutti, A.; Bachmann, F.; Faivre, D. Biohybrid and Bioinspired Magnetic Microswimmers. Small 2018, 14, 1704374. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, R.; Rybenkov, V.V. A Guide to Magnetic Tweezers and Their Applications. Front. Phys. 2016, 4. [Google Scholar] [CrossRef]
- Sitters, G.; Kamsma, D.; Thalhammer, G.; Ritsch-Marte, M.; Peterman, E.J.G.; Wuite, G.J.L. Acoustic force spectroscopy. Nat. Methods 2015, 12, 47–50. [Google Scholar] [CrossRef]
- Shishkin, I.; Markovich, H.; Roichman, Y.; Ginzburg, P. Auxiliary Optomechanical Tools for 3D Cell Manipulation. Micromachines 2020, 11, 90. [Google Scholar] [CrossRef] [Green Version]
- Hayakawa, T.; Fukada, S.; Arai, F. Fabrication of an On-Chip Nanorobot Integrating Functional Nanomaterials for Single-Cell Punctures. IEEE Trans. Robot. 2014, 30, 59–67. [Google Scholar] [CrossRef]
- Nawaz, S.; Sanchez, P.; Bodensiek, K.; Li, S.; Simons, M.; Schaap, I.A.T. Cell Visco-Elasticity Measured with AFM and Optical Trapping at Sub-Micrometer Deformations. PLoS ONE 2012, 7, e45297. [Google Scholar] [CrossRef]
- Vargas-Pinto, R.; Gong, H.; Vahabikashi, A.; Johnson, M. The Effect of the Endothelial Cell Cortex on Atomic Force Microscopy Measurements. Biophys. J. 2013, 105, 300–309. [Google Scholar] [CrossRef] [Green Version]
- Dy, M.C.; Kanaya, S.; Sugiura, T. Localized cell stiffness measurement using axial movement of an optically trapped microparticle. J. Biomed. Opt. 2013, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndoye, F.; Yousafzai, M.S.; Coceano, G.; Bonin, S.; Scoles, G.; Ka, O.; Niemela, J.; Cojoc, D. The influence of lateral forces on the cell stiffness measurement by optical tweezers vertical indentation. Int. J. Optomechatron. 2016, 10, 53–62. [Google Scholar] [CrossRef]
- Yousafzai, M.S.; Coceano, G.; Mariutti, A.; Ndoye, F.; Amin, L.; Niemela, J.; Bonin, S.; Scoles, G.; Cojoc, D. Effect of neighboring cells on cell stiffness measured by optical tweezers indentation. J. Biomed. Opt. 2016, 21, 57004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayano, S.; Wakamoto, Y.; Yamashita, S.; Yasuda, K. Quantitative measurement of damage caused by 1064-nm wavelength optical trapping of Escherichia coli cells using on-chip single cell cultivation system. Biochem. Biophys. Res. Commun. 2006, 350, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Vu, K.T.; Krishnan, P.; Trang, T.C.; Shin, D.; Kimel, S.; Berns, M.W. Wavelength dependence of cell cloning efficiency after optical trapping. Biophys. J. 1996, 70, 1529–1533. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, M.B.; Oddershede, L.B.; Siegumfeldt, H. Optical tweezers cause physiological damage to Escherichia coli and Listeria bacteria. Appl. Environ. Microbiol. 2008, 74, 2441–2446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vizsnyiczai, G.; Kelemen, L.; Ormos, P. Holographic multi-focus 3D two-photon polymerization with real-time calculated holograms. Opt. Express 2014, 22, 24217–24223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuman, K.C.; Block, S.M. Optical trapping. Rev. Sci. Instrum. 2004, 75, 2787–2809. [Google Scholar] [CrossRef]
- Ohashi, T.; Ishii, Y.; Ishikawa, Y.; Matsumoto, T.; Sato, M. Experimental and numerical analyses of local mechanical properties measured by atomic force microscopy for sheared endothelial cells. Bio-Med. Mater. Eng. 2002, 12, 319–327. [Google Scholar]
- Pesen, D.; Hoh, J.H. Micromechanical Architecture of the Endothelial Cell Cortex. Biophys. J. 2005, 88, 670–679. [Google Scholar] [CrossRef] [Green Version]
- Feneberg, W.; Aepfelbacher, M.; Sackmann, E. Microviscoelasticity of the Apical Cell Surface of Human Umbilical Vein Endothelial Cells (HUVEC) within Confluent Monolayers. Biophys. J. 2004, 87, 1338–1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathur, A.B.; Collinsworth, A.M.; Reichert, W.M.; Kraus, W.E.; Truskey, G.A. Endothelial, cardiac muscle and skeletal muscle exhibit different viscous and elastic properties as determined by atomic force microscopy. J. Biomech. 2001, 34, 1545–1553. [Google Scholar] [CrossRef]
- Coceano, G.; Yousafzai, M.S.; Ma, W.; Ndoye, F.; Venturelli, L.; Hussain, I.; Bonin, S.; Niemela, J.; Scoles, G.; Cojoc, D.; et al. Investigation into local cell mechanics by atomic force microscopy mapping and optical tweezer vertical indentation. Nanotechnology 2016, 27, 065102. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.R.; Charras, G.T. Experimental validation of atomic force microscopy-based cell elasticity measurements. Nanotechnology 2011, 22, 345102. [Google Scholar] [CrossRef]
- Chiou, Y.W.; Lin, H.K.; Tang, M.J.; Lin, H.H.; Yeh, M.L. The Influence of Physical and Physiological Cues on Atomic Force Microscopy-Based Cell Stiffness Assessment. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grexa, I.; Fekete, T.; Molnár, J.; Molnár, K.; Vizsnyiczai, G.; Ormos, P.; Kelemen, L. Single-Cell Elasticity Measurement with an Optically Actuated Microrobot. Micromachines 2020, 11, 882. https://doi.org/10.3390/mi11090882
Grexa I, Fekete T, Molnár J, Molnár K, Vizsnyiczai G, Ormos P, Kelemen L. Single-Cell Elasticity Measurement with an Optically Actuated Microrobot. Micromachines. 2020; 11(9):882. https://doi.org/10.3390/mi11090882
Chicago/Turabian StyleGrexa, István, Tamás Fekete, Judit Molnár, Kinga Molnár, Gaszton Vizsnyiczai, Pál Ormos, and Lóránd Kelemen. 2020. "Single-Cell Elasticity Measurement with an Optically Actuated Microrobot" Micromachines 11, no. 9: 882. https://doi.org/10.3390/mi11090882