Abstract
Main conclusion
Specific and common genes including transcription factors, resistance genes and pathways were significantly induced in potato by Phytophthora infestans, Ralstonia solanacearum, and Potato virus Y infection.
Abstract
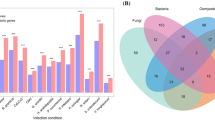
The three major pathogens, namely, Phytophthora infestans, Ralstonia solanacearum, and Potato virus Y, can cause late blight, bacterial wilt, and necrotic ringspot, respectively, and thus severely reduce the yield and quality of potatoes (Solanum tuberosum L.). This study was the first to systematically analyze the relationship between transcriptome alterations in potato infected by these pathogens at the early stages. A total of 75,500 unigenes were identified, and 44,008 were annotated into 5 databases, namely, non-redundant (NR), Swiss-Prot protein, clusters of orthologous groups for eukaryotic complete genomes (KOG), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. A total of 6945 resistance genes and 11,878 transcription factors (TFs) were identified from all transcriptome data. Differential expression analysis revealed that 13,032 (9490 specifics), 9877 (6423 specifics), and 6661 (4144 specifics) differentially expressed genes (DEGs) were generated from comparisons of the P. infestans/control (Pi vs. Pi-CK), R. solanacearum/control (Rs vs. Rs-CK), and PVY/control (PVY vs. PVY-CK) treatments, respectively. The specific DEGs from the 3 comparisons were assigned to 13 common pathways, such as biosynthesis of amino acids, plant hormone signal transduction, carbon metabolism, and starch and sucrose metabolism. Weighted Gene Co-Expression Network Analysis (WGCNA) identified many hub unigenes, of which several unigenes were reported to regulate plant immune responses, such as FLAGELLIN-SENSITIVE 2 and chitinases. The present study provide crucial systems-level insights into the relationship between transcriptome changes in potato infected with the three pathogens. Moreover, this study presents a theoretical basis for breeding broad-spectrum and specific pathogen-resistant cultivars.





Similar content being viewed by others
Abbreviations
- DEGs:
-
Differentially expressed genes
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- PVY:
-
Potato virus Y
- R genes:
-
Resistance genes
- TFs:
-
Transcript factors
- WGCNA:
-
Weighted Gene Co-Expression Network Analysis
References
Adachi H, Nakano T, Miyagawa N, Ishihama N, Yoshioka M, Katou Y, Yaeno T, Shirasu K, Yoshioka H (2015) WRKY transcription tactors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27:2645–2663. https://doi.org/10.1105/tpc.15.00213
Ali MC, Karasev AV (2015) Potato mosaic and tuber necrosis. In: Tennant P, Fermin G (eds) Virus diseases of tropical and subtropical crops. CABI, Boston, pp 94–107
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/s0022-2836(05)80360-2
Amorim LLB, Santos DFD (2017) Transcription factors involved in plant resistance to pathogens. Curr Protein Pept Sci 18:335–351. https://doi.org/10.2174/1389203717666160619185308
Bari R, Jones JD (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69:473–488. https://doi.org/10.1007/s11103-008-9435-0
Birch PRJ, Bryan G, Fenton B, Gilroy EM, Toth IK (2012) Crops that feed the world 8: Potato: Are the trends of increased global production sustainable? Food Secur 4:477–508. https://doi.org/10.1007/s12571-012-0220-1
Cantwell JD (2017) A great-grandfather's account of the Irish potato famine (1845–1850). Baylor Univ Med Center 30:382–383. https://doi.org/10.1080/08998280.2017.11929657
Chen L, Guo X, Xie C, He L, Cai X, Tian L, Song B, Liu J (2013) Nuclear and cytoplasmic genome components of Solanum tuberosum + S. chacoense somatic hybrids and three SSR alleles related to bacterial wilt resistance. Theor Appl Genet 126:1861–1872. https://doi.org/10.1007/s00122-013-2098-5
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY (2014) CytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8(Suppl 4):S11. https://doi.org/10.1186/1752-0509-8-s4-s11
Cletus J, Balasubramanian V, Vashisht D, Sakthivel N (2013) Transgenic expression of plant chitinases to enhance disease resistance. Biotechnol Lett 35:1719–1732. https://doi.org/10.1007/s10529-013-1269-4
Collonnier C, Mulya K, Fock II, Mariska II, Servaes A, Vedel F, Siljak-Yakovlev S, Souvannavong VV, Ducreux G, Sihachakr D (2001) Source of resistance against Ralstonia solanacearum in fertile somatic hybrids of eggplant (Solanum melongena L.) with Solanum aethiopicum L. Plant Sci 160:301–313. https://doi.org/10.1016/s0168-9452(00)00394-0
Cruz AP, Ferreira V, Pianzzola MJ, Siri MI, Coll NS, Valls M (2014) A novel, sensitive method to evaluate potato germplasm for bacterial wilt resistance using a luminescent Ralstonia solanacearum reporter strain. Mol Plant Microbe Interact 27:277–285. https://doi.org/10.1094/mpmi-10-13-0303-fi
Doncheva NT, Morris JH, Gorodkin J, Jensen LJ (2019) Cytoscape StringApp: Cytoscape StringApp: Network analysis and visualization of proteomics data. J Proteome Res 18:623–632. https://doi.org/10.1021/acs.jproteome.8b00702
Duan Y, Duan S, Armstrong MR, Xu J, Zheng J, Hu J, Chen X, Hein I, Li G, Jin L (2020) Comparative transcriptome profiling reveals compatible and tncompatible patterns of potato toward Phytophthora infestans. Bethesda 10:623–634. https://doi.org/10.1534/g3.119.400818
Fass MI, Rivarola M, Ehrenbolger GF, Maringolo CA, Montecchia JF, Quiroz F, García-García F, Blázquez JD, Hopp HE, Heinz RA, Paniego NB, Lia VV (2020) Exploring sunflower responses to Sclerotinia head rot at early stages of infection using RNA-seq analysis. Sci Rep 10:13347. https://doi.org/10.1038/s41598-020-70315-4
Fock II, Collonnier C, Purwito A, Luisetti J, Souvannavong VV, Vedel F, Servaes A, Ambroise A, Kodja H, Ducreux G, Sihachakr D (2000) Resistance to bacterial wilt in somatic hybrids between Solanum tuberosum and Solanum phureja. Plant Sci 160:165–176. https://doi.org/10.1016/s0168-9452(00)00375-7
French ER, Anguiz R, Aley P (1998) The usefulness of potato Rrsistance to Ralstonia solanacearum, for the integrated control of bacterial wilt. In: Prior P, Allen C, Elphinstone J (eds) Bacterial wilt disease. Springer, Berlin, pp 381–385
Fry WE, Birch PR, Judelson HS, Grunwald NJ, Danies G, Everts KL, Gevens AJ, Gugino BK, Johnson DA, Johnson SB, McGrath MT, Myers KL, Ristaino JB, Roberts PD, Secor G, Smart CD (2015) Five reasons to consider Phytophthora infestans a reemerging pathogen. Phytopathology 105:966–981. https://doi.org/10.1094/phyto-01-15-0005-fi
Gong L, Han S, Yuan M, Ma X, Hagan A, He G (2020) Transcriptomic analyses reveal the expression and regulation of genes associated with resistance to early leaf spot in peanut. BMC Res Notes 13:381. https://doi.org/10.1186/s13104-020-05225-9
Goyer A, Hamlin L, Crosslin JM, Buchanan A, Chang JH (2015) RNA-Seq analysis of resistant and susceptible potato varieties during the early stages of potato virus Y infection. BMC Genomics 16:472. https://doi.org/10.1186/s12864-015-1666-2
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Gutierrez Sanchez PA, Babujee L, Jaramillo Mesa H, Arcibal E, Gannon M, Halterman D, Jahn M, Jiang J, Rakotondrafara AM (2020) Overexpression of a modified eIF4E regulates potato virus Y resistance at the transcriptional level in potato. BMC Genomics 21:18. https://doi.org/10.1186/s12864-019-6423-5
Hane DC, Hamm PB (1999) Effects of seedborne potato virus Y Infection in two potato cultivars expressing mild disease symptoms. Plant Dis 83:43–45. https://doi.org/10.1094/pdis.1999.83.1.43
Ishihama N, Yoshioka H (2012) Post-translational regulation of WRKY transcription factors in plant immunity. Curr Opin Plant Biol 15:431–437. https://doi.org/10.1016/j.pbi.2012.02.003
Jiang G, Wei Z, Xu J, Chen H, Zhang Y, She X, Macho AP, Ding W, Liao B (2017) Bacterial wilt in China: History, current status, and future perspectives. Front Plant Sci. https://doi.org/10.3389/fpls.2017.01549
Kangasjarvi S, Tikkanen M, Durian G, Aro EM (2014) Photosynthetic light reactions–an adjustable hub in basic production and plant immunity signaling. Plant Physiol Biochem 81:128–134. https://doi.org/10.1016/j.plaphy.2013.12.004
Karasev AV, Gray SM (2013) Continuous and emerging challenges of Potato virus Y in potato. Annu Rev Phytopathol 51:571–586. https://doi.org/10.1146/annurev-phyto-082712-102332
Kawa D (2020) Security notice: This plant immunity is under mRNA surveillance. Plant Cell 32:803–804. https://doi.org/10.1105/tpc.20.00169
Kim-Lee H, Moon JS, Hong Y, Kim MS, Cho HM (2005) Bacterial wilt resistance in the progenies of the fusion hybrids between haploid of potato and Solanum commersonii. Amer J Potato Res 82:129–137
Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, SjoBack R, SjoGreen BR, Strombom L (2006) The real-time polymerase chain reaction. Mol Aspects Med 27:95–125
Lacomme C, Glais L, Bellstedt DU, Dupuis B, Karasev AV, Jacquot E (eds) (2017) Potato virus Y: biodiversity, pathogenicity, epidemiology and management. Publ, Springer Intern
Lagaert S, Belien T, Volckaert G (2009) Plant cell walls: Protecting the barrier from degradation by microbial enzymes. Semin Cell Dev Biol 20:1064–1073. https://doi.org/10.1016/j.semcdb.2009.05.008
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. https://doi.org/10.1186/1471-2105-9-559
Lemaga B, Kakuhenzire R, Kassa B, Ewell PT, Priou S (2005) Integrated control of potato bacterial wilt in Eastern Africa: The experience of African highlands initiative. In: Allen C, Prior P, Hayward AC (eds) Bacterial wilt disease and the Ralstonia solanacearum species complex. American Phytopathological Society Press, St Paul, MN, pp 145–158
Liu Q, Liu Y, Tang Y, Chen J, Ding W (2017) Overexpression of NtWRKY50 increases resistance to Ralstonia solanacearum and alters salicylic acid and jasmonic acid production in tobacco. Front Plant Sci 8:1710. https://doi.org/10.3389/fpls.2017.01710
Liu Y, Liu Q, Tang Y, Ding W (2019) NtPR1a regulates resistance to Ralstonia solanacearum in Nicotiana tabacum via activating the defense-related genes. Biochem Biophys Res Commun 508:940–945. https://doi.org/10.1016/j.bbrc.2018.12.017
Mabuchi K, Maki H, Itaya T, Suzuki T, Nomoto M, Sakaoka S, Morikami A, Higashiyama T, Tada Y, Busch W, Tsukagoshi H (2018) MYB30 links ROS signaling, root cell elongation, and plant immune responses. Proc Natl Acad Sci USA 115:e4710–e4719. https://doi.org/10.1073/pnas.1804233115
Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629. https://doi.org/10.1111/j.1364-3703.2012.00804.x
Mehle N, Gutierrez-Aguirre I, Prezelj N, Delic D, Vidic U, Ravnikar M (2014) Survival and transmission of potato virus Y, pepino mosaic virus, and potato spindle tuber viroid in water. Appl Environ Microbiol 80:1455–1462. https://doi.org/10.1128/aem.03349-13
Meng F, Yao J, Allen C (2011) A MotN mutant of Ralstonia solanacearum is hypermotile and has reduced virulence. J Bacteriol 193:2477–2486. https://doi.org/10.1128/jb.01360-10
Mizubuti ESG, Fry WE (2006) Potato late blight. In: Cooke B, Jones D, Kaye B (eds) The epidemiology of plant diseases. Springer, Dordrecht, pp 445–471
Mou S, Liu Z, Gao F, Yang S, Su M, Shen L, Wu Y, He S (2017) CaHDZ27, a homeodomain-leucine zipper I protein, positively regulates the resistance to Ralstonia solanacearum infection in pepper. Mol Plant Microbe Interact 30:960–973. https://doi.org/10.1094/mpmi-06-17-0130-r
Nakashima K, Yamaguchi-Shinozaki K (2013) ABA signaling in stress-response and seed development. Plant Cell Rep 32:959–970. https://doi.org/10.1007/s00299-013-1418-1
Naseem M, Srivastava M, Tehseen M, Ahmed N (2015) Auxin crosstalk to plant immune networks: a plant-pathogen interaction perspective. Curr Protein Pept Sci 16:389–394. https://doi.org/10.2174/1389203716666150330124911
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27:29–34. https://doi.org/10.1093/nar/27.1.29
Orosa B, Yates G, Verma V, Srivastava AK, Srivastava M, Campanaro A, De Vega D, Fernandes A, Zhang C, Lee J, Bennett MJ, Sadanandom A (2018) SUMO conjugation to the pattern recognition receptor FLS2 triggers intracellular signalling in plant innate immunity. Nat Commun 9:5185. https://doi.org/10.1038/s41467-018-07696-8
Otulak K, Garbaczewska G (2010) Localisation of hydrogen peroxide accumulation during Solanum tuberosum cv. Rywal hypersensitive response to Potato virus Y. Micron 41:327–335. https://doi.org/10.1016/j.micron.2009.12.004
Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7:e30619. https://doi.org/10.1371/journal.pone.0030619
Peeters N, Guidot A, Vailleau F, Valls M (2013) Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol Plant Pathol 14:651–662. https://doi.org/10.1111/mpp.12038
Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B, Tsai J, Quackenbush J (2003) TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19:651–652. https://doi.org/10.1093/bioinformatics/btg034
Raffaele S, Farrer RA, Cano LM, Studholme DJ, MacLean D, Thines M, Jiang RH, Zody MC, Kunjeti SG, Donofrio NM, Meyers BC, Nusbaum C, Kamoun S (2010) Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science (New York, NY) 330:1540–1543. https://doi.org/10.1126/science.1193070
Ren H, Gray WM (2015) SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol Plant 8:1153–1164. https://doi.org/10.1016/j.molp.2015.05.003
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Schonfeld J, Heuer H, Van Elsas JD, Smalla K (2003) Specific and sensitive detection of Ralstonia solanacearum in soil on the basis of PCR amplification of fliC fragments. Appl Environ Microbiol 69:7248–7256. https://doi.org/10.1128/aem.69.12.7248-7256.2003
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. https://doi.org/10.1038/nprot.2012.016
Valkonen JP (2015) Elucidation of virus-host interactions to enhance resistance breeding for control of virus diseases in potato. Breed Sci 65:69–76. https://doi.org/10.1270/jsbbs.65.69
Verma V, Ravindran P, Kumar PP (2016) Plant hormone-mediated regulation of stress responses. BMC Plant Biol 16:86. https://doi.org/10.1186/s12870-016-0771-y
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63. https://doi.org/10.1038/nrg2484
Wang X, Boevink P, McLellan H, Armstrong M, Bukharova T, Qin Z, Birch PR (2015) A host KH RNA-binding protein is a susceptibility factor targeted by an RXLR effector to promote late blight disease. Mol Plant 8:1385–1395. https://doi.org/10.1016/j.molp.2015.04.012
Wang J, Gao C, Li L, Cao W, Chu Z (2019) Transgenic RXLR effector PITG_15718.2 suppresses immunity and reduces vegetative growth in potato. Int J Mol Sci 20:30–31. https://doi.org/10.3390/ijms20123031
Whisson SC, Boevink PC, Wang S, Birch PR (2016) The cell biology of late blight disease. Curr Opin Microbiol 34:127–135. https://doi.org/10.1016/j.mib.2016.09.002
Whitworth JL, Nolte P, McIntosh C, Davidson R (2006) Effect of Potato virus Y on yield of three potato cultivars grown under different nitrogen levels. Plant Dis 90:73–76. https://doi.org/10.1094/pd-90-0073
Yu J, Ai G, Shen D, Chai C, Jia Y, Liu W, Dou D (2019) Bioinformatical analysis and prediction of Nicotiana benthamiana bHLH transcription factors in Phytophthora parasitica resistance. Genomics 111:473–482. https://doi.org/10.1016/j.ygeno.2018.03.005
Yue C, Cao H, Lin H, Hu J, Ye Y, Li J, Hao Z, Hao X, Sun Y, Yang Y, Wang X (2019) Expression patterns of alpha-amylase and beta-amylase genes provide insights into the molecular mechanisms underlying the responses of tea plants (Camellia sinensis) to stress and postharvest processing treatments. Planta 250:281–298. https://doi.org/10.1007/s00425-019-03171-w
Zhang C, Chen H, Cai T, Deng Y, Zhuang R, Zhang N, Zeng Y, Zheng Y, Tang R, Pan R, Zhuang W (2017) Overexpression of a novel peanut NBS-LRR gene AhRRS5 enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Biotechnol J 15:39–55. https://doi.org/10.1111/pbi.12589
Zhao Y (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61:49–64. https://doi.org/10.1146/annurev-arplant-042809-112308
Zhu S, Li Y, Vossen JH, Visser RG, Jacobsen E (2012) Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Res 21:89–99. https://doi.org/10.1007/s11248-011-9510-1
Zuluaga AP, Sole M, Lu H, Gongora-Castillo E, Vaillancourt B, Coll N, Buell CR, Valls M (2015) Transcriptome responses to Ralstonia solanacearum infection in the roots of the wild potato Solanum commersonii. BMC Genomics 16:246. https://doi.org/10.1186/s12864-015-1460-1
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31720103912) and Shandong“Double Tops” Program (SYL2017XTTD11).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cao, W., Gan, L., Shang, K. et al. Global transcriptome analyses reveal the molecular signatures in the early response of potato (Solanum tuberosum L.) to Phytophthora infestans, Ralstonia solanacearum, and Potato virus Y infection. Planta 252, 57 (2020). https://doi.org/10.1007/s00425-020-03471-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-020-03471-6