Pathogenicity of Shiga Toxin Type 2e Escherichia coli in Pig Colibacillosis
- 1Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia Romagna “Bruno Ubertini,” Brescia, Italy
- 2Department of Veterinary Public Health and Food Safety, Istituto Superiore di Sanità, Rome, Italy
Shiga toxin type 2e (Stx2e) Escherichia coli is the causative factor of diarrhea and edema in swine. The aims of this study were to determine the prevalence of Stx2e-producing E. coli isolates and to characterize isolates from clinical cases of pig colibacillosis and healthy swine. During the 11 years of the study (2006–2017), a total of 233 Stx2e-producing isolates were detected−230 out of 2,060 (11.16%) E. coli isolated from diseased pigs and 3 out of 171 (1.75%) from healthy swine. Stx2e-producing isolates were indeed more present in clinical colibacillosis cases than in healthy pigs (p = 0.0002). The predominant serogroup was O139 (79.82%) and the most common fimbrial factor present in these isolates was F18 (177 isolates), followed by F6 (5 isolates). The enterotoxins LTI, STa, and STb were detected in 10.43, 41.73, and 48.26% of the isolates, respectively. The predominant virotypes F18-Stx2e and -STa-STb-Stx2e were similarly present in weaners (33.33 and 35.52%) and finishers (38.30 and 25.53%). Among isolates from diseased pigs, O139 and F18 were the more frequently identified serogroup and virulence factor, respectively. Of the tested 230 Stx2e-producing isolates isolated from diseased pigs, 29 (12.60%) harbored genes encoding ESBL, particularly TEM (79.30%), CTX-M1 (17.20%), and CMY-2 (3.40%). Antimicrobial resistance to tetracycline was the most common characteristic (98.25%), followed by ampicillin (93.91%), cephalotin (90.43%) and trimethoprim/sulfamethoxazole (82.17%). Our results showed that Stx2e-producing E. coli were more frequently associated with clinical forms of colibacillosis, with minimal probability to isolate these isolates from healthy pigs.
Introduction
Escherichia coli is a facultative anaerobe (1) and, although an opportunistic pathogen commonly found in the intestinal tracts of vertebrates (2), has the potential to cause seriously pathogenic colibacillosis in humans and animals, when it harbors specific virulence genes (3). Indeed, even if most E. coli are harmless, some of them are able to cause gastroenteric/enteric or systemic diseases in vertebrates (4, 5). According to Barbau-Piednoir et al. (6), pathogenic E. coli are divided in diarrhoeagenic E. coli (DEC) and extraintestinal E. coli (ExPEC) and, based on the type of virulence factor present and on the host clinical symptoms, these groups can be further grouped into pathotypes. Uropathogenic (UPEC) and neonatal meningitidis (NMEC) E. coli belong to the ExPEC while the DEC group consists of eight pathotypes: shigatoxigenic (STEC) [including the enterohemorrhagic (EHEC)], enteropathogenic (EPEC), enterotoxigenic (ETEC), enteroinvasive (EIEC), enteroaggregative (EAggEC), diffusively adherent (DAEC), adherent invasive (AIEC), and the recently described enteroaggregative shigatoxigenic (EAggSTEC) E. coli (6).
Focusing on intensive pig farming, colibacillosis is a major threat due to severe economic losses, consequences of high morbidity, increased mortality rates, stunted or decreased growth, elevated health management costs and elevated costs of pharmacological treatments (7, 8). Among pathogenic E. coli, ETEC, and STEC isolates are the main agents in swine causing post-weaning diarrhea (PWD) and edema disease (ED), respectively (9). A common feature of these two pathotypes is the expression of specific fimbrial adhesins that allow the bacterial binding at the mucosal surface of the porcine small intestine, contrasting intestinal peristalsis (5). These fimbriae, indicated with the letter F, are numbered progressively. In swine, isolates typically display specific types of fimbriae, including F4 (K88), F5 (K99), F6 (P987), F18, and F41 (10). The most commonly reported fimbrial adhesins are of the F4 and F18 types, both with different antigenic variants: three for F4 (ab, ac, and ad), with F4ac being the most prevalent, and two main variants for F18 (F18ab is associated with ED and F18ac with PWD (11)). Other associated fimbriae of lower prevalence include F5, F6, and F41, whose number of active receptors on the intestinal epithelial cells decreases with the age of the host (5, 10). In addition to these colonization factors, the pathogenic attitude of ETEC and STEC is also mediated by the ability to produce enterotoxins and/or Shiga-toxins (12), respectively. The effect of both toxins on the digestive system leads to rapid intestinal fluid hypersecretion with consequent sudden onset of osmotic watery diarrhea that frequently results in severe dehydration and circulatory shock (13). The heat-labile enterotoxin LT and the heat-stable enterotoxins STa and STb are the best-known toxins (8). Epithelial adherence of these toxins is predominantly facilitated by adhesive fimbriae, thus according to Renzhammer et al. (8), the detection of at least one enterotoxin gene, together with one gene coding for fimbriae (F4, F5, F6, F18, and F41) in a single E. coli is defined as an essential criterion for the classification of porcine ETEC. On the other hand, STEC (Shiga toxin-producing E. coli) isolates produce the Shiga-toxins that are classified as type 1 (Stx1: Shiga-toxin 1 type) with subtypes a, c, d, and as type 2 (Stx2: Shiga-toxin 2 type) with subtypes a, b, c, d, e, f and g. Stx2 variant “e,” also called edemigenic toxin, is the causative agent of the severe ED in pigs (9). The ability of STEC to cause disease is related to the production of one or more Shiga-like toxins, which inhibits the protein synthesis of host cells, thus leading to cell death (14). Furthermore, some isolates harbor both the Stx2e and enterotoxin genes, being able to cause symptoms of edema disease and diarrhea in the same animal (STEC/ETEC) (15). Therefore, the fact that ED is almost the most pathogenic among pig colibacillosis, together with the attitude of STEC in general to be important food-borne pathogens (7), representing a serious zoonotic risk with swine playing an important role as a carrier (13), highlights the need to investigate the presence and the characteristics of these pathogenic isolates in pigs.
In order to provide further insights into patterns associated with virulent and non-virulent phenotypes of Stx2e, the present study aims to determine the prevalence, the biomolecular and antimicrobial resistance patterns of Stx2e-producing E. coli isolates isolated from cases of pig colibacillosis and from healthy swine.
Materials and Methods
Sample Collection
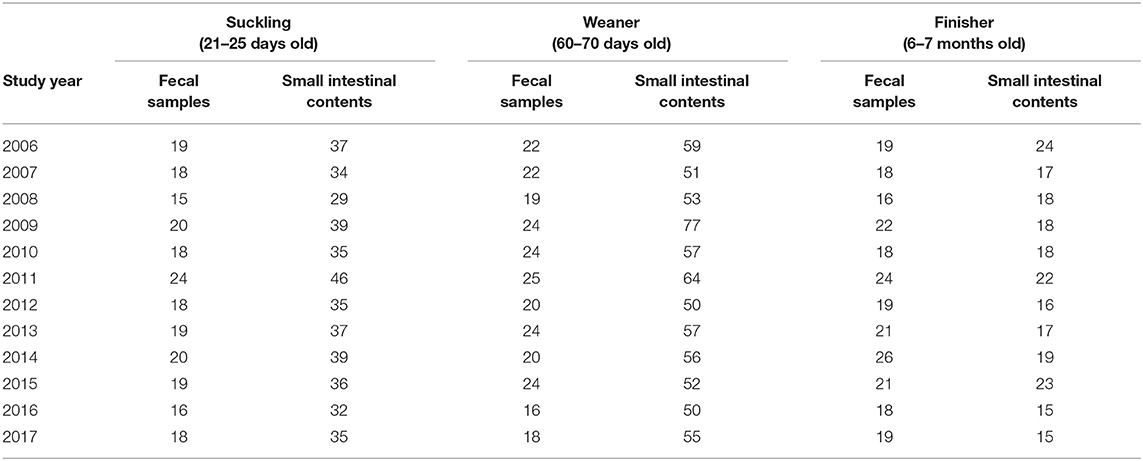
The presence of Stx2e-producing E. coli isolates was investigated in 2060 cases of colibacillosis, from January 2006 to December 2017, as part of the routine activity of the Diagnostic Section of IZSLER in Brescia, Italy The presence of Stx2e-producing E. coli isolates was investigated in 2060 cases of colibacillosis, from January 2006 to December 2017, as part of the routine activity of the Diagnostic Section of IZSLER in Brescia, Italy. In particular, a total of 1,337 E. coli were isolated from small intestinal contents sampled during the necropsy of pigs (Table 1) conferred to our Department for further diagnostic investigations. At the macroscopic examination these pigs showed yellowish, gray, or slightly pink watery diarrhea with a characteristic smell; the small intestine were dilatated, slightly edematous and hyperaemic with enlarged and hyperaemic mesenteric lymph nodes. Moreover, edema disease occurred in 24 of these cases and were characterized by edema in various sites, e.g., eye lids, nose bridge and forehead, stomach wall, and the mesentery of the colon, as main finds of edema disease as Franke et al. (16) had previously reported. In addition, an overall 723 E. coli were isolated from fecal samples/fecal swabs of living pigs (Table 1) with diarrhea. Carcasses and fecal samples were conferred to our Department from a total of 670 farms (no more than five samples from the same farm during the whole study period). All of these farms make a regular use of antibiotics.
The presence of Stx2e-producing E. coli isolates was also investigated in 171 living pigs−29 weaners and 142 finishers—without clinical any signs of colibacillosis sampled from 18 distinct farms during 2016. Farmers were voluntarily involved in the study and who agreed to participate allowed us to collect feces and/or fecal swabs from their swine.
This study was carried out as a part of the routine activity of Diagnostic Section of Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia Romagna (IZSLER), thus the scientific protocol did not require an additional approval of the Ethical Committee for Animal Experimentation of IZSLER.
Identification of E. coli
The isolation procedure was consistent during the whole study period (2006–2017). The samples, processed within 24 h after the collection, were cultured on MacConkey agar plates and blood agar plates (Oxoid, Italy) and incubated aerobically for 18 ± 2 h at 37 ± 2°C. After an overnight incubation, suspicious E. coli colonies were identified by morphology (pink on MacConkey and/or with hemolysis on blood agar plates) and Gram staining. For each case/animal, one suspected colony with presumptive biochemical properties (lactose and indole positive; H2S, oxidase, and urease negative) was subcultured on BHI (Brain Heart Infusion) agar slant (Oxoid, Italy) while its identity was confirmed by the biochemical method, API 20E (bioMérieux, France).
All the isolates were preserved in medium containing tryptone soy broth (TSB) with 20% glycerol at −80°C.
Serogrouping of the Isolates
The serogrouping of the Stx2e-producing E. coli isolated from clinical cases of colibacillosis was based on somatic O-antigens since, among them, there are the most frequent and pathogenic for humans and animals (17). The analyses was carried out using agglutinating antisera in microplate according to Guinée et al. (18) and modified by Blanco et al. (19). The available antisera were against 30 serogroups from O1 to O157 (O1, O2, O5, O8, O9, O15, O18, O20, O22, O26, O45, O49, O55, O64, O78, O86, O88, O101, O103, O111, O113, O118, O128, O138, O139, O141, O147, O149, O153, O157) (Oxoid, Italy) were tested. Briefly, in 96-well plates, 100 μl of diluted antiserum and 100 μl of O-antigens suspensions prepared by heating bacterial suspensions for 1 h at 100°C were mixed into each well and incubated at 37 ± 2°C for 18 ± 2 h. A positive reaction was confirmed by agglutination in the diluted antiserum. Isolates that did not react with any of the O-antisera examined were classified as O-antisera untypeable (ND).
Molecular Characterization by Multiplex Real-Time PCR
Escherichia coli isolated from both clinical cases and healthy pigs were screened by multiplex PCR for the presence of the major virulence genes of porcine pathogenic E. coli, including genes for 5 different adhesins (K88, K99, F41, 987P, and F18) and 4 different toxins (LT, STaP, STb, and Stx2e) following the method according to Casey and Bosworth (20). Briefly, DNA was obtained from each E. coli isolate (one colony) by hot lysis procedure in which the sample were harvested by centrifugation (12.000 × g for 5 min), and washed three times in distilled water, boiled at 97.5 ± 2.5°C for 10 min and immediately cooled on ice for 2 min. After centrifugation, the extracted DNA was subjected to multiplex PCR for screening of virulence factors (VFs) using specific primers (Table 2). According to Casey and Bosworth (20), the PCR reaction mixtures contained 18 primers at a concentration of 0.5 μmol each, 0.2 mmol deoxyribonucleotide triphosphate mix, 1 X reaction buffer, 5 mmol MgCl2, and 2.5 units of Taq polymerase in a final volume of 20 μl. These amplification conditions were followed: an initial denaturation at 94°C for 10 min, followed by 30 cycles of denaturation for 30 s at 94°C, annealing at 55°C for 45 s, and extension for 1.5 min at 72°C. The extension time was increased by 3 s each cycle, and the final extension was 10 min at 72°C. The amplification products were then separated and detected by electrophoresis using 4% agarose gels at 75–100 V for 1.5–2 h (20).
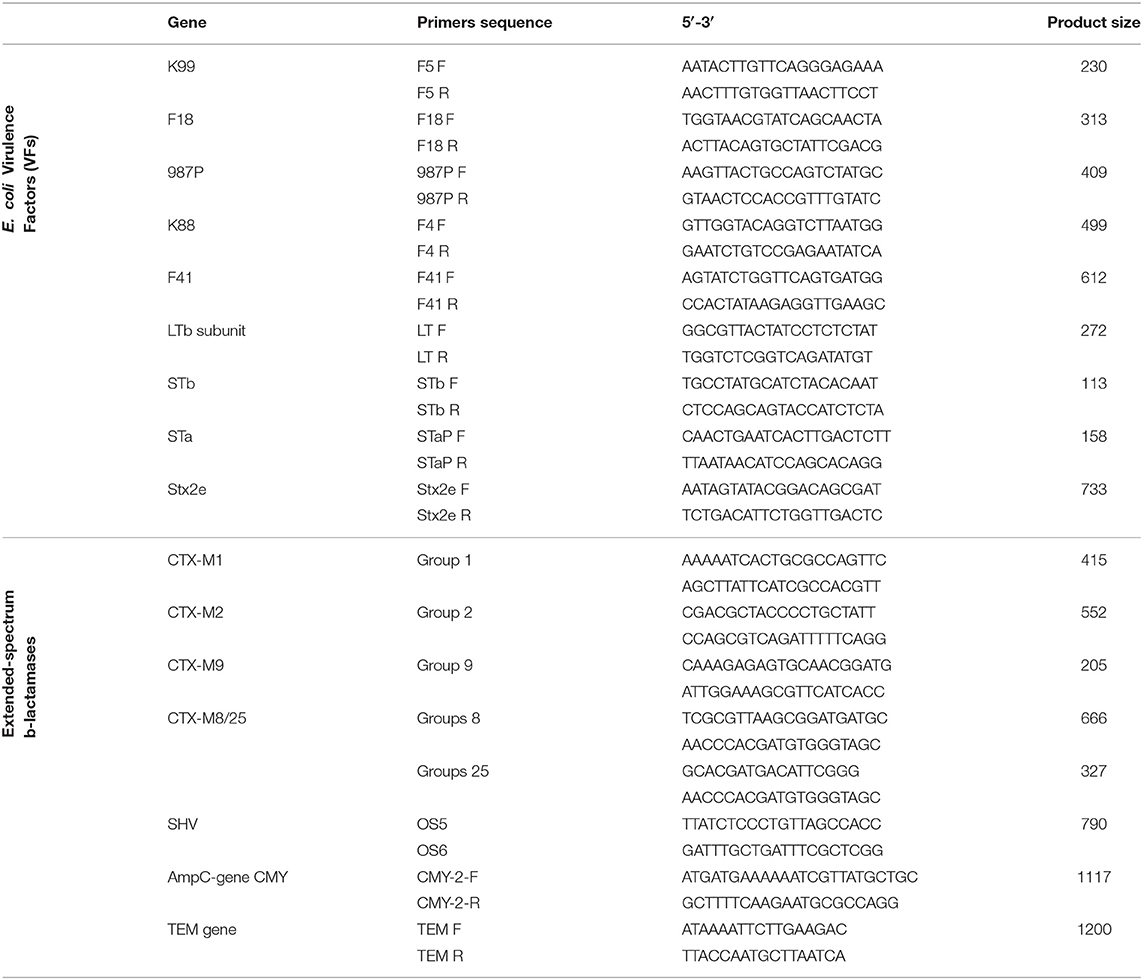
Table 2. PCR primers used for the detection of E. coli virulence factors (VFs) and extended-spectrum β-lactamases (ESBL).
Antimicrobial Susceptibility Testing
The susceptibility of Stx2e-producing isolates to a panel of antimicrobials was tested using the disc diffusion method following the procedures of the Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute (CLSI) (21–26). Briefly, the isolates were inoculated in trypticase soy broth (TSB) and then plated on Mueller-Hinton agar. The following 11 commercially available antibiotic discs were used: ampicillin (AMP: 10 μg), amoxicillin/clavulanic acid (AMC: 30 μg), cephalothin (KF: 30 μg), ceftiofur (EFT: 30 μg), enrofloxacin (ENR: 5 μg), florfenicol (FFC: 30 μg), flumequine (FQ: 30 μg), gentamicin (G: 10 μg), kanamycin (K: 30 μg), tetracycline (TE: 30 μg), trimethoprim/sulfamethoxazole (SXT: 1.25/23.75 μg). The plates were read after incubation in aerobic condition at 37 ± 2°C to 18 ± 2 h. The isolates were classified as resistant, susceptible or intermediate to the antimicrobials tested according to the zone diameter interpretative standard recommendations by CLSI (M45-A; M2-A9; M100-S26; VET08; M100-S29). The isolates with intermediate growth were considered to be resistant (27).
The identification of broad spectrum β-lactamase -producing E. coli was performed through a double synergy diagnostic method: after the pre-enrichment with BHI broth supplemented with 1 mg/L cefotaxime and an overnight incubation, a drop of the BHI broth was used to inoculate MacConkey agar supplemented with 1 mg/L cefotaxime (28–30). Positive growths were identified as pink to dark-pink colonies and one of these was selected for further molecular characterization.
Characterization of β-Lactamase Genes
Detection of the resistance genes present in the Stx2e-producing isolates isolated from diseased pigs was performed using a panel of PCR reactions. A multiplex PCR was used for the identification of the CTX-M group genes whose single or multiple positivity identifies the five main phylogenetic groups—CTX-M1, CTX-M2, CTX-M9 and CTX-M8 and CTX-M25 (31). In addition, single PCR reactions were used for the identification of the SHV gene (32), TEM gene (33), using universal primers as previously described (34–37), and AmpC genes (CMY-2, CMY-4, CMY-6, CMY-7, CMY-12, CMY-13, CMY-14, CMY-18, LAT-3) [(38); Table 2]. All the TEM and SHV PCR amplicons were DNA-sequenced.
Statistical Analysis
Comparison between groups was assessed by using Fisher's exact test and differences were considered significant when P < 0.05.
Results
Sample Identification and Molecular Characterization
A total of the 2,060 E. coli were isolated from diseased pigs. Of these, 230 (11.16%) were positive for Stx2e-producing isolates (Table 3), β-hemolytic activity was recorded in 215 (93.48%) of the isolates. All the details about the Stx2e-producing E. coli isolated from diseased pigs during each study year are available in Table S1. Three out of 171 (1.75%) E. coli isolated from healthy pigs harbored the Stx2e toxin (Table 3). Stx2e-producing isolates were more present in clinical colibacillosis cases than in healthy pigs (p = 0.0002).
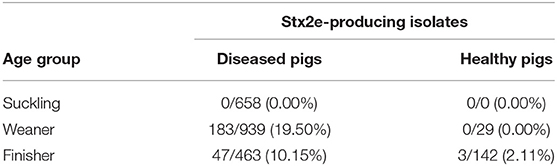
Table 3. Proportion of Stx2e-producing isolates E. coli isolated in diseased and healthy weaners and finishers pigs.
Serogrouping of the Isolates
The serogrouping of the 230 Stx2e-producing E. coli isolated from diseased pigs showed that 109 (47.39%) of them were typeable with available O-antisera, whereas 121 (52.61%) isolates could not be assigned to any of the 30 serotypes serogroups tested and defined as O-antisera untypeable (ND). A total of 11 serogroups were identified within the collection: O1 (0.92%, 1/109), O2 (2.75%, 3/109), O6 (0.92%, 1/109), O8 (2.75%, 3/109), O9 (0.92%, 1/109), O128 (1.83%, 2/109), O139 (79.82%, 87/109), O141 (4.59%, 5/109), O147 (2.75%, 3/109), O149 (0.92%, 1/109), O157 (1.83%, 2/109).
Virulence Factors (VFs) Detected in Stx2e-Producing Isolates
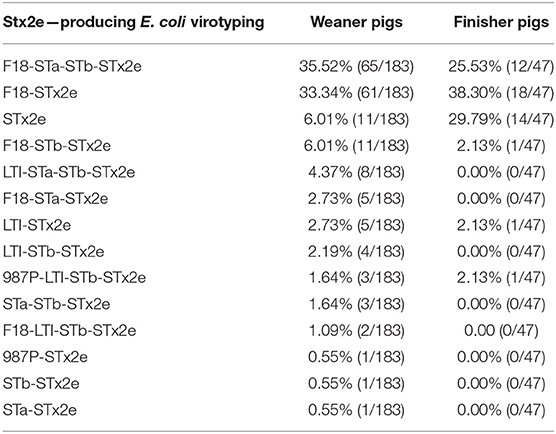
Fimbrial factors were detected in 182 (79.13%) of the 230 Stx2e-producing isolates isolated from clinical colibacillosis cases. The most common fimbrial factor in the isolates was F18 (177 isolates) followed by 987P (5 isolates) (Figure 1). Differences in the presence of fimbrial factors between weaners and finishers are shown in Table 4. In particular, F18 was more frequently found in weaner than in finishers (p < 0.05). The enterotoxin LTI was detected in 24 out of 230 (10.43%) isolates; STa was detected in 96 out of 230 (41.73%) Stx2e-producing isolates; while STb was detected in 111 out of 230 (48.26%) Stx2e-producing isolates recorded (Figure 1). In addition, the prevalence of predominant virotypes, F18-Stx2e and F18-STa-STb-Stx2e, was similarly present in weaners (33.33 and 35.52%) and finishers (38.30 and 25.53%) (Table 4). No virulence factors were detected in the 3 Stx2e-producing E. coli isolated from healthy pigs.
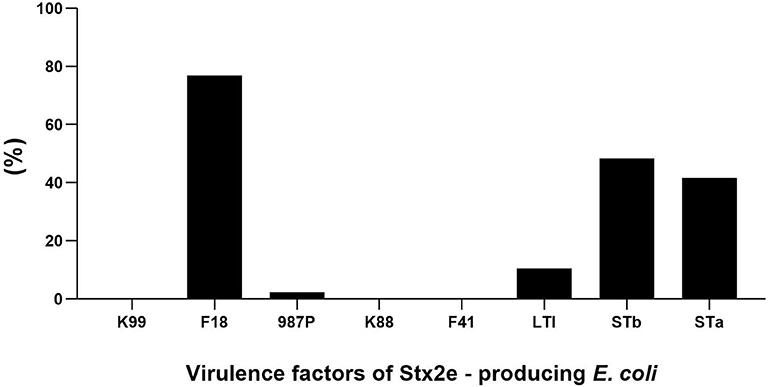
Figure 1. Distribution of fimbrial factors and enterotoxins in the overall analyzed 230 Stx2e-producing isolates.
Antimicrobial Susceptibility Testing
The antimicrobials resistance profile of the Stx2e-producing E. coli isolated from diseased pigs was determined against 11 antibiotics (Table 5). The whole panel of antimicrobials had been applied on 189 Stx2e-producing isolates, for which frequency of multi-drug resistance (MDR) was calculated (Table 5), since not all the considered antibiotics have been available during all the study period. All these isolates were resistant to at least one antibiotic (Tables 5, 6). Resistance to tetracycline had the highest prevalence with a rate of 98.25%; ampicillin resistance showed a rate of 93.91% and resistance to cephalotin a rate of 90.43% (Table 5). Other common resistances possessed by the Stx2e-producing isolates detected in clinical colibacillosis cases were to trimethoprim/sulfamethoxazole, kanamycin, gentamicin, followed by resistance to amoxicillin/clavulanic acid, flumequine, ceftiofur, florfenicol, and enrofloxacin (Table 5). The overall possible multi-drug resistant profiles of the Stx2e-producing E. coli for which the whole panel of antimicrobials has been applied are available in Table S2. Out of the 230 Stx2e-producing isolates recorded from diseased pigs, 29 (12.60%) harbored genes encoding ESBL (Figure 2). The ESBL genes identified were TEM (79.30%), CTX-M1 (17.20%), and CMY-2 (3.40%) (Figure 2). From the total of 23 Stx2e-producing E. coli TEM-positive, overall 21 of them were available for sequencing and showed TEM-1 as result. PCR products were sequenced and deposited in NCBI GenBank with accession numbers from MT789713 to MT789733.
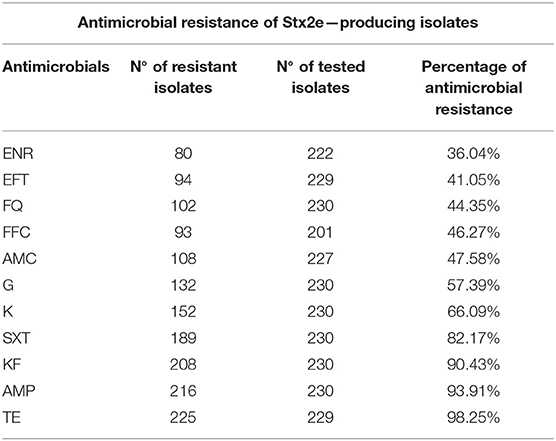
Table 5. List of antimicrobials included in the Kirby-Bauer disc diffusion method, Number (N°) of resistant and tested Stx2e-producing isolates and the percentages of antimicrobial resistance identified in this study.
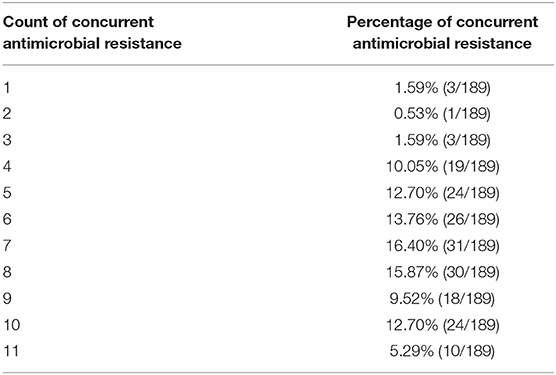
Table 6. Distribution of concurrent resistance in Stx2e-producing E. coli isolated from diseased pigs.
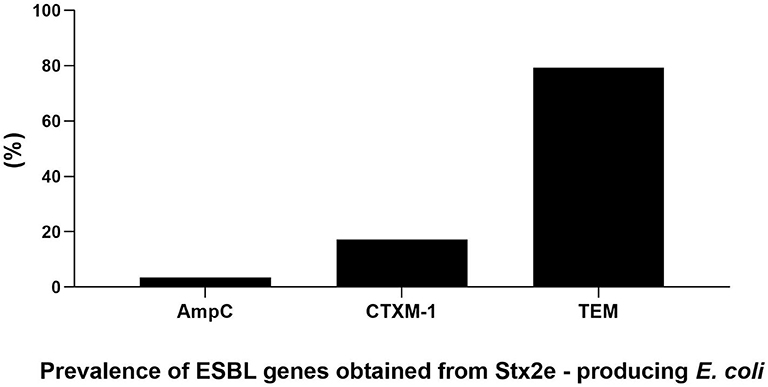
Figure 2. Prevalence and genetic characterization of ESBL genes obtained from the overall 230 Stx2e-producing isolates isolated from diseased pigs.
Discussion
In the present study, Stx2e-producing E. coli were isolated and characterized from diseased and healthy weaner and finisher pigs, in light of the considerably different distribution and frequency of serogroups and virotypes between production stages (10, 19). A total of 230 isolates obtained from clinical forms of colibacillosis showed the Shiga-toxin variant 2 (Stx2e). Only three cases of Stx2e-producing isolates were isolated from healthy pigs. Thus, the Stx2e variant appears to be more frequently associated with clinical colibacillosis. The O139 serogroup and the virulence factor F18 were isolated in Stx2e-producing isolates from diseased pigs, suggesting that these characteristics are associated with a virulent phenotype.
The presence of Stx2e-producing isolates in cases of colibacillosis in pigs is well-documented although our study recorded higher prevalence than previously reported. For instance, Chen et al. (39) pointed out that Stx2e-producing isolates were isolated in 6.1% of the 250 E. coli isolates collected from pigs with diarrhea. A similar prevalence was reported by Brand et al. (40), who found 3.36% of Stx2e-producers in E. coli isolates from 115 pigs with diarrhea. Moreover, the presence of Stx2e-producing E. coli in healthy pigs has not been well-investigated although these isolates were normally isolated from healthy swine (41). Meng et al. found the presence of these subtype isolates in 25.42% of healthy swine in slaughterhouses (13). Similar result emerged in a recent study by Arancia et al., which showed a prevalence of 25.8% for Stx2e subtype isolates (52.1%) in the caecal contents of slaughtered pigs (41). Compared to these studies, we observed a lower prevalence of Stx2e subtype isolates in healthy pigs. This finding could be ascribed to possible differences in the screening methods employed. In the work by Meng et al. (13), the reported rate of 25.42% of STEC in healthy pigs was obtained by PCR screening; however, only 6.18% of the swine samples yielded STEC isolates by microbiological culture. Moreover, the difference in reported prevalence of isolation could be due to the anatomic sites of sampling. Indeed, the rate of isolation of STEC in fecal samples was considerably lower than the rate from colon or the small intestine (13). As this regards, the fact of having sampled feces/fecal swabs of healthy pigs and the known presence in animal feces of numerous materials (e.g., complex polysaccharides, bilirubin, and bile salts) that are inhibitors of PCR (42) should be considered in the interpretation of results, although the reliability of both the used DNA extraction and PCR protocols.
We found that the vast majority of the isolated Stx2e-producing isolates belong to the O139 serogroup. Interestingly, available studies have mainly focused on the distribution of O157, without consideration for other serogroups. Milnes et al. (43) reported the detection of 0.3% (6/2,000) of VTEC O157 isolates from swine fecal samples at slaughter facilities. Similarly, Lenahan et al. (44) recorded a prevalence of STEC O157:H7 of 0.6% (3/480) in fecal samples from swine at slaughterhouses, supporting the report by Bonardi et al. (45) who isolated a low proportion (0.7%, 1/150) of O157-STEC isolates from fecal samples at slaughter facilities. Several European studies have reported STEC prevalence in swine population and most focused on STEC serogroup O157 (39, 46). However, in the present study, O157 was isolated in only one case out of the 230 investigated. Further, Friedrich et al. (47) showed that pigs with edema disease in post-weaning and young finishing pigs caused by STEC presented with serogroups O8, O138, O139, O141, and O147, similar to findings on Stx2e isolates reported by Fratamico et al. (14). Overall, these data suggest that, although O157 seemed an important serogroup from a public health perspective, other serogroups can have a significant impact on animal health, in certain types of animals (7).
In our study, most of the Stx2e-producing isolates harbored F18 adhesin factor, with a lower percentage of samples possessing 987P fimbriae; K88, K99, and F41 were not observed. These findings are not surprising, as the gene encoding F18 fimbriae is among the most important ones associated with colibacillosis and edema disease (10). Likely the fact that all the Stx2e-producing E. coli were detected from diseased animals with diarrhea supports an effect of the co-presence of these two virulence factors (Stx2e toxin and F18 adhesin) in causing this symptom, although even Shiga toxigenic E. coli are reported to cause diarrhea in young pigs (48–50). F18 fimbriae are absent in most human-derived STEC, but are essential for adherence to swine epithelial cells (13). Chen et al. (39) reported that 50.2% E. coli isolates detected from pigs with post-weaning diarrhea carried one or more fimbrial factors. In the present study, data from the multiplex PCR on Stx2e-producing isolates showed that STa and STb positive isolates were more prevalent than LTI. This fact, in light of the attitude of these enterotoxins in causing diarrhea and dehydration in pigs (51), may be related to the diarrheic syndrome recorded in the most of the analyzed subjects. In addition, this finding agrees with previous studies by Chen et al. (39) that reported 60.5% STa in isolates from pigs with post-weaning diarrhea, and Toledo et al. (52), which observed STa (30%), STb (17%), Stx2e (6%), and LTI (5%) as the most common enterotoxins in suckling and weaning pigs.
The presence of extended-spectrum β-lactamase producing Escherichia coli (ESBL) in humans and animals is a major global public health concern (53). In the present study, 29 out of the 230 Stx2e–producing isolates (12.60%) isolated from diseased pigs possessed ESBL genes. Although STEC isolates are not deemed a reservoir of ESBL, in recent reports have indicated an association between STEC isolates and ESBL genes, Mandakini et al. (53) reported on 25.29% isolates phenotypically confirmed as ESBL producers. We found that the predominant genes in our ESBL isolates were resistance to TEM (79.30%), CTX-M1 (17.20%), and CMY-2 (3.40%), in contrast to Jones et al. (54) who pointed out that CTX-M resistant genes were detected in 85% of isolates while TEM resistance was found in 6.12%. However, although the phenotypical test carried out in our study demonstrated the resistance of these isolates to cefotaxime, the fact that sequencing analyses showed the presence of TEM-1 Stx2e–producing E. coli cannot exclude the possibility that these isolates could have any other susceptibility to other cephalosporins (55).
Antimicrobial resistance (AMR) has become a major public health concern because bacteria causing infectious diseases are becoming less susceptible to antibiotic treatment (56, 57). However, antibiotic resistance can also arise in opportunistic bacteria, such as E. coli, as a result of different mechanisms (56). Currently, the presence and the dissemination of AMR among the genus Escherichia spp. represents an emerging problem (56, 57). In the present study, Stx2e-producing isolates isolated from farmed pigs showed high levels of resistance to various antimicrobial agents. Although the use of the Minimum Inhibitory Concentration (MIC) test is currently suggested for antimicrobial resistance detection, we employed the Kirby-Bauer disc-diffusion method because specimen from 11 years of study were included, with results dating back to 2006 when MIC was not routinely performed. The highest percentages of antimicrobial resistance recorded for Stx2e-producing strains isolated in this study were registered for tetracycline (98.25%), ampicillin (93.91%), cefalotin (90.43%), and trimethoprim/sulfamethoxazole (82.17%). These findings appeared not related to a specific correlation between Stx2e toxins and phenotypical antimicrobial resistance but these high percentages could be ascribed to the wide use of these antibiotics in the past for treating pig respiratory and enteric bacterial diseases (27). Moreover, the high percentage of isolates resistant to antibiotics commonly used to treat diarrhea in pigs emphasizes the importance of avoiding unnecessary use of antibiotics. The observed high level of resistance to tetracycline and ampicillin is probably a direct consequence of the intense use of these antibiotics in veterinary medicine. In addition, the horizontal transfer of these resistant genes should be considered together with their potential acquisition from a contaminated external environment (58, 59). We found that, besides the high rate of resistance to most of the tested antimicrobials, the majority of Stx2e-producing isolates isolated from pigs also exhibited multidrug resistance. This data agrees with results from a study by Brand et al. (40) that found a high level of AMR, in particular to tetracycline (50%), sulfamethoxazole (49%), trimethoprim (34%), and ampicillin (26%) in E. coli isolates from pigs. These results and the increasing awareness on drug-resistant E. coli strains, related to the prophylactic use of antibiotics on healthy piglets or as food additive (60), highlight the need to develop a vaccine against Stx2e-producing isolates as an useful method alternatively to antimicrobials (61). In addition, an effective vaccine program is desirable in order to prime immunity able to confirm protection to the effect of toxins (60).
In conclusion, the present study contributes to strengthening available information on the virulence factors of Stx2e-producing isolates in diseased and healthy pigs. In addition, this survey sheds new light on potential pathogenic characteristics of Stx2e-producing isolates and points out the need for a rational management of antibiotics use and effective vaccination programs in swine farms in order to minimize the impact of these antimicrobial resistant pathogenic Stx2e-producing E. coli isolates on animal health.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
Ethical review and approval was not required for the animal study because this study was carried out as a part of the routine activity of Diagnostic Section of Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia Romagna (IZSLER), thus the scientific protocol did not require an additional approval of the Ethical Committee for Animal Experimentation of IZSLER.
Author Contributions
VB, CS, SG, PP, and GA contributed to the conception and design of the study. VB, LB, AP, MD'I, MB, and AG performed the experiments. VB and LB organized the database. VB performed the statistical analysis and wrote the first draft of the manuscript. NF and MB wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This research was supported by in house funding of IZSLER and by the Italian Ministry of Health (IZSLER project PRC 2014/001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Federico Scali for his valuable contribution that improved the manuscript and Alberto Tiraboschi for his technical support with the sampling activities. We thank Editage Group (www.editage.com) for editing a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.545818/full#supplementary-material
References
1. Milton AAP, Agarwal RK, Priya GB, Aravind M, Athira CK, Rose L, et al. Captive wildlife from India as carriers of Shiga toxin-producing, enteropathogenic and enterotoxigenic Escherichia coli. J Vet Med Sci. (2019) 81:321–7. doi: 10.1292/jvms.18-0488
2. Khairy RM, Mohamed ES, Ghany HMA, Abdelrahim SS. Phylogenic classification and virulence genes profiles of uropathogenic E. coli and diarrhegenic E. coli strains isolated from community acquired infections. PLoS ONE. (2019) 14:e0222441. doi: 10.1371/journal.pone.0222441
3. Hayer SS, Rovira A, Olsen K, Johnson TJ, Vannucci F, Rendahl A, et al. Prevalence and trend analysis of antimicrobial resistance in clinical Escherichia coli isolates collected from diseased pigs in the USA between 2006 and 2016. Transbound Emerg Dis. (2020) 1–12. doi: 10.1111/tbed.13528. [Epub ahead of print].
4. Center for Disease Control and Prevention (CDC). Foodborne Disease Active Surveillance Network (FoodNet). Atlanta, GA (2016)
5. Luppi A. Swine enteric colibacillosis: diagnosis, therapy and antimicrobial resistance. Porcine Health Manag. (2017) 3:16. doi: 10.1186/s40813-017-0063-4
6. Barbau-Piednoir E, Denayer S, Botteldoorn N, Dierick K, De Keersmaecker SCJ, Roosens NH. Detection and discrimination of five E. coli pathotypes using a combinatory SYBR® Green qPCR screening system. Appl Microbiol Biotechnol. (2018) 102:3267–85. doi: 10.1007/s00253-018-8820-0
7. Tseng M, Fratamico PM, Bagi L, Delannoy S, Fach P, Manning SD, et al. Molecular characterization of Shiga toxin-producing E. coli (STEC) from finishing swine in a longitudinal study. Appl Environ Microbiol. (2014) 80:6395–402. doi: 10.1128/AEM.01761-14
8. Renzhammer R, Loncaric I, Roch F-F, Pinior B, Käsbohrer A, Spergser J, et al. Prevalence of virulence genes and antimicrobial resistances in E. coli associated with neonatal diarrhea, postweaning diarrhea, and edema disease in pigs from Austria. Antibiotics. (2020) 9:208. doi: 10.3390/antibiotics9040208
9. Scheutz F, Teel LD, Beutin L, Piérard D, Buvens G, Karch H, et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol. (2012) 50:2951–63. doi: 10.1128/JCM.00860-12
10. Vu-Khac H, Holoda E, Pilipcinec E, Blanco M, Blanco JE, Dahbi G, et al. Serotypes. virulence genes, intimintypes and PFGE profiles of Escherichia coli isolated from piglets with diarrhoea in Slovakia. Vet J. (2007) 174:176–87. doi: 10.1016/j.tvjl.2006.05.019
11. Francis DH. Enterotoxigenic Escherichia coli infection in pigs and its diagnosis. J Swine Health Prod. (2002) 10:171–5. doi: 10.1155/2018/6434910
12. Zhang W, Mellmann A, Sonntag A, Wieler L, Bielaszewska M, Tschäpec H, et al. Structural and functional differences between disease-associated genes of enterohaemorrhagic Escherichia coli O111. Int J Med Microbiol. (2007) 297:17–26. doi: 10.1016/j.ijmm.2006.10.004
13. Meng Q, Bai X, Zhao A, Lan R, Du H, Wang T, et al. Characterization of Shiga toxin-producing Escherichia coli isolated from healthy pigs in China. BMC Microbiol. (2014) 14:5. doi: 10.1186/1471-2180-14-5
14. Fratamico PM, Bagi LK, Bush EJ, Solow BT. Prevalence and characterization of Shiga toxin-producing Escherichia coli in Swine feces recovered in the National Animal Health Monitoring System's Swine 2000 study. Appl Environ Microbiol. (2004) 70:7173–8. doi: 10.1128/AEM.70.12.7173–7178.2004
15. Barth S, Schwanitz A, Bauerfeind R. Polymerase chain reaction-based method for the typing of F18 fimbriae and distribution of F18 fimbrial subtypes among porcine Shiga toxin–encoding Escherichia coli in Germany. J Vet Diagn Investig. (2011) 23:454–64. doi: 10.1177/1040638711403417
16. Franke S, Gunzer F, Wieler LH, Baljer G, Karch H. Construction of recombinant Shiga-like toxin-IIv (SLT-11~) and its use in monitoring the SLT-11~antibody status of pigs. Vet Microbiol. (1995) 43:41–52.
17. Szczerba-Turek A, Socha P, Bancerz-Kisiel A, Platt-Samoraj A, Lipczynska-Ilczuk K, Siemionek J, et al. Pathogenic potential to humans of Shiga toxin-producing Escherichia coli isolated from wild boars in Poland. Int J Food Microbiol. (2019) 300:8–13. doi: 10.1016/j.ijfoodmicro.2019.04.004
18. Guinée PAM, Agterberg CM, Jansen WH. Escherichia coli 0 antigen typing by means of a mechanized microtechnique. Appl Microbiol. (1972) 24:127–31.
19. Blanco M, Blanco JE, Gonzalez EA, Mora A, Jansen W, Gomes TAT, et al. Genes coding for enterotoxins and verotoxins in porcine Escherichia coli isolates belonging to different O:K:H serotypes: relationship with toxic phenotypes. J Clin Microbiol. (1997) 35:2958–63.
20. Casey TA, Bosworth BT. Design and evaluation of a multiplex polymerase chain reaction assay for the simultaneous identification of genes for nine different virulence factors associated with Escherichia coli that cause diarrhea and edema disease in swine. J Vet Diagn Invest. (2009) 21:25–30. doi: 10.1177/104063870902100104
21. Clinical and Laboratory Standards Institute (CLSI). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; Approved Guideline (M45-A). Wayne, PA (2006).
22. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard. 9th ed (M2-A9). Wayne, PA (2006).
23. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. Approved Standard. 3rd ed. Wayne, PA: CLSI document M31-A3 (2008).
24. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. Wayne, PA: CLSI Supplement M100-S26 (2016).
25. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. 4th ed. Wayne, PA: CLSI supplement VET08 (2018).
26. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. Wayne, A: CLSI supplement M100 (2019).
27. Luppi A, Bonilauri P, Dottori M, Gherpelli Y, Biasi G, Merialdi G, et al. Antimicrobial resistance of F4+ Escherichia coli isolated from swine in Italy. Transbound Emerg Dis. (2015) 62:67–71. doi: 10.1111/tbed.12081
28. Wasyl D, Hasman H, Cavaco LM, Aarestrup FM. Prevalence and characterization of cephalosporin resistance in nonpathogenic Escherichia coli from food-producing animals slaughtered in Poland. Microb Drug Resist. (2012) 18:79–82. doi: 10.1089/mdr.2011.0033
29. Cavaco L, Hendriksen RS, Agersø Y, Svendsen CA, Nielsen H, Guerra B, et al. Selective enrichment of ESBL, AmpC and carbapenemase producing E. coli in meat and cecal samples additional validation for poultry samples. In: Abstract from 26th European Congress o Clinical Microbiology and Infectious diseases (ECCMID 2016). Amsterdam (2016).
30. Van Damme I, Garcia-Graells C, Biasino W, Gowda T, Botteldoorn N, De Zutter L. High abundance and diversity of extended-spectrum beta-lactamase (ESBL) producing Escherichia coli in faeces and tonsils of pigs at slaughter. Vet Microbiol. (2017) 208:190–4. doi: 10.1016/j.vetmic.2017.08.009
31. Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J Antimicrob Chemother. (2006) 57:154–5. doi: 10.1093/jac/dki412
32. Arlet G, Rouveau M, Philippon A. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum β-lactamase. FEMS Microbiol Lett. (1997) 152:163–67. doi: 10.1111/j.1574-6968.1997.tb10423.x
33. Mabilat C, Goussard S, Sougakoff W, Spencer RC, Courvalin P. Direct sequencing of the amplified structural gene and promoter for the extended-broad-spectrum β-lactamase TEM-9 (RHH-1) of Klebsiella pneumonia. Plasmid. (1990) 23:27–34. doi: 10.1016/0147-619x(90)90041-a
34. Chang F-Y, Siu LK, Fung C-P, Huang M-H, Ho M. Diversity of SHV and TEM b-lactamases in klebsiella pneumoniae: gene evolution in northern Taiwan and two novel b-lactamases, SHV-25 and SHV-26. Antimicrob Agents Chemother. (2001) 45:2407–13. doi: 10.1128/AAC.45.9.2407–2413.2001
35. Ahmed AM, Motoi Y, Sato M, Maruyama A, Watanabe H, Fukumoto Y, et al. Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl Environ Microbiol. (2007) 73:6686–90. doi: 10.1128/AEM.01054-07
36. Crémet L, Caroff N, Dauvergne S, Reynaud A, Lepelletier D, Corvec S. Prevalence of plasmid-mediated quinolone resistance determinants in ESBL enterobacteriaceae clinical isolates over a 1-year period in a French hospital. Pathol Biol. (2011) 59:151–6.
37. Gbonon V, Afran SA, Guessennd KN, Toty AA, Diplo TFB, N'Guetta ASP, et al. Detection of TEM and SHV genes in clinical Escherichia coli and Klebsiella pneumoniae strains ESBL isolated in neonatology and pediatric units. Int J Microbiol Res. (2018) 23:1–7. doi: 10.9734/MRJI/2018/39354
38. Dierikx C, van Essen-Zandbergen A, Veldman K, Smith H, Mevius D. Increased detection of extended-spectrum β-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet Microbiol. (2010) 145:273–8. doi: 10.1016/j.vetmic.2010.03.019
39. Chen X, Gao S, Jiao X, Fan Liu X. Prevalence of serogroups and virulence factors of Escherichia coli isolates isolated from pigs with postweaning diarrhoea in eastern China. Vet Microbiol. (2004) 103:13–20. doi: 10.1016/j.vetmic.2004.06.014
40. Brand P, Gobeli S, Perreten V. Pathotyping and antibiotic resistance of porcine enterovirulent Escherichia coli isolates from Switzerland (2014-2015). Schweiz Arch Tierheilkd. (2017) 159:373–80. doi: 10.17236/sat00120
41. Arancia S, Iurescia M, Lorenzetti S, Stravino F, Buccella C, Caprioli A, et al. Detection and isolation of Shiga toxin-producing Escherichia coli (STEC) isolates in caecal samples from pigs at slaughter in Italy. Vet Med Sci. (2019) 5:462–9 doi: 10.1002/vms3.175
42. Fratamico PM, Bagi LK, Pepe T. A multiplex polymerase chain reaction assay for rapid detection and identification of Escherichia coli O157:H7 in foods and bovine feces. J Food Prot. (2000) 63:1032–7. doi: 10.4315/0362-028x-63.8.1032
43. Milnes AS, Sayers AR, Stewart I, Clifton-Hadley FA, Davies RH, Newell DG, et al. Factors related to the carriage of Verocytotoxigenic E. coli, Salmonella, thermophilic Campylobacter and Yersinia enterocolitica in cattle, sheep and pigs at slaughter. Epidemiol Infect. (2009) 137:1135–48. doi: 10.1017/S095026880900199X
44. Lenahan M, O'Brien SB, Byrne C, Ryan M, Kennedy CA, McNamara EB, et al. Molecular characterization of Irish E. coli O157: H7 isolates of human, bovine, ovine and porcine origin. J Appl Microbiol. (2009) 107:1340–9. doi: 10.1111/j.1365-2672.2009.04320.x
45. Bonardi S, Brindani F, Pizzin G, Lucidi L, D'Incau M, Liebana E, et al. Detection of Salmonella spp., Yersinia enterocolitica and verocytotoxin-producing Escherichia coli O157 in pigs at slaughter in Italy. Int J Food Microbiol. (2003) 85:101–10. doi: 10.1016/s0168-1605(02)00504-4
46. Feder I, Morgan Wallace F, Gray JT, Fratamico P, Fedorka-Cray PJ, Pearce RA, et al. Isolation of Escherichia coli O157:H7 from intact colon faecal samples of swine. Emerg Infect Dis. (2003) 9:380–3. doi: 10.3201/eid0903.020350
47. Friedrich AW, Bielaszewska M, Zhang WL, Pulz M, Kuczius T, Ammon A, et al. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J Infect Dis. (2002) 185:74–84. doi: 10.1086/338115
48. Liu W, Yuan C, Meng X, Du Y, Gao R, Tang J, et al. Frequency of virulence factors in Escherichia coli isolated from suckling pigs with diarrhoea in China. Vet J. (2014) 199:286–9. doi: 10.1016/j.tvjl.2013.11.019
49. Osek J. Virulence factors and genetic relatedness of Escherichia coli strains isolated from pigs with post-weaning diarrhea. Vet Microbiol. (2000) 71:211–22. doi: 10.1016/s0378-1135(99)00168-6
50. Osek J. Genetic diversity among Escherichia coli O149:K91 strains isolated from pigs with diarrhoea determined by randomly amplified polymorphic DNA analysis. Res Vet Sci. (1999) 67:195–6. doi: 10.1053/rvsc.1999.0285
51. Kwon D, Choi C, Jung T, Chung H-K, Kim J-R, Bae S-S, et al. Genotypic prevalence of the fimbrial adhesins (F4, F5, F6, F41 and F18) and toxins (LT, STa, STb and Sbc2e) in Escherichia coli isolated from postweaning pigs with diarrhoea or oedema disease in Korea. Vet Rec. (2002) 150:35–7. doi: 10.1056/NEJMoa1716153
52. Toledo A, Gomez D, Cruz C, Carreon R, Lopez J, Giono S, et al. Prevalence of virulence genes in Escherichia coli isolates isolated from piglets in the suckling and weaning period in Mexico. J Med Microbiol. (2012) 61:148–56. doi: 10.1099/jmm.0.031302-0
53. Mandakini R, Dutta TK, Chingtham S, Roychoudhury P, Samanta I, Joardar SN, et al. ESBL-producing Shiga-toxigenix E. coli (STEC) associated with piglet diarrhea in India. Trop Anim Health Prod. (2015) 47:377–81. doi: 10.1007/s11250-014-0731-1
54. Jones CH, Tuckman M, Keeney D, Ruzin A, Bradford PA. Characterization and sequence analysis of extended-spectrum-β-lactamase-encoding genes from Escherichia coli, Klebsiella pneumonia, and Proteus mirabilis isolates collected during tigecycline phase 3 clinical trials. Antimicrob Agents Chemother. (2009) 53:465–75. doi: 10.1128/AAC.00883-08
55. Doi Y, Iovleva A, Bonomo RA. The ecology of extended-spectrum b-lactamases (ESBLs) in the developed world. J Travel Med. (2017) 24(Suppl. 1):S44–51 doi: 10.1093/jtm/taw102
56. Smith MG, Jordan D, Chapman TA, Chin JJ, Barton MD, Do TN, et al. Antimicrobial resistance and virulence gene profiles in multi-drug resistant enterotoxigenic Escherichia coli isolated from pigs with post-weaning diarrhoea. Vet Microbiol. (2010) 145:299–307. doi: 10.1016/j.vetmic.2010.04.004
57. Yang H, Paruch L, Chen X, van Eerde A, Skomedal H, Wang Y, et al. Antibiotic application and resistance in Swine production in China: current situation and future perspectives. Front Vet Sci. (2019) 6:136. doi: 10.3389/fvets.2019.00136
58. Mohan Raj JR, Vittal R, Shivakumaraswamy SK, Deekshit VK, Chakraborty A, Karunasagar I. Presence & mobility of antimicrobial resistance in Gram-negative bacteria from environmental samples in coastal Karnataka, India. Indian J Med Res. (2019) 149:290–4. doi: 10.4103/ijmr.IJMR_2088_17
59. Fouz N, Pangesti KNA, Yasir M, Al-Malki AL, Azhar EI, Hill-Cawthorne GA, et al. The contribution of wastewater to the transmission of antimicrobial resistance in the environment: implications of mass gathering settings. Trop Med Infect Dis. (2020) 5:33. doi: 10.3390/tropicalmed5010033
60. Hamabata T, Sato T, Takita E, Matsui T, Imaoka T, Nakanishi N, et al. Shiga toxin 2eB-transgenic lettuce vaccine is effective in protecting weaned piglets from edema disease caused by Shiga toxin-producing Escherichia coli infection. Anim Sci J. (2019) 90:1460–7. doi: 10.1111/asj.13292
Keywords: edema disease, healthy pigs, virulence factors, antimicrobial resistance, ESBL genes
Citation: Baldo V, Salogni C, Giovannini S, D'Incau M, Boniotti MB, Birbes L, Pitozzi A, Formenti N, Grassi A, Pasquali P and Alborali GL (2020) Pathogenicity of Shiga Toxin Type 2e Escherichia coli in Pig Colibacillosis. Front. Vet. Sci. 7:545818. doi: 10.3389/fvets.2020.545818
Received: 26 March 2020; Accepted: 12 August 2020;
Published: 18 September 2020.
Edited by:
Joy Scaria, South Dakota State University, United StatesReviewed by:
Indranil Samanta, West Bengal University of Animal and Fishery Sciences, IndiaMagaly Toro, University of Chile, Chile
Copyright © 2020 Baldo, Salogni, Giovannini, D'Incau, Boniotti, Birbes, Pitozzi, Formenti, Grassi, Pasquali and Alborali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicoletta Formenti, nicoletta_formenti@yahoo.it
 Valentina Baldo1
Valentina Baldo1  Mario D'Incau
Mario D'Incau Maria Beatrice Boniotti
Maria Beatrice Boniotti Nicoletta Formenti
Nicoletta Formenti Paolo Pasquali
Paolo Pasquali Giovanni Loris Alborali
Giovanni Loris Alborali