Recent and Rapid Radiation of the Highly Endangered Harlequin Frogs (Atelopus) into Central America Inferred from Mitochondrial DNA Sequences
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Molecular Laboratory Protocols
2.3. Phylogenetic Analyses
2.4. Biogeographic Analyses
2.5. Pairwise Genetic Distances
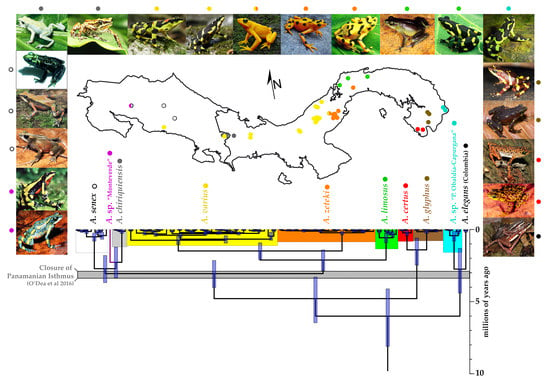
3. Results
3.1. Phylogenetic Analyses
3.2. Divergence Times and Ancestral Area Estimation
| Node Label | Mean Crown Age (Ma) | 95% Posterior Credibility Interval | DEC Estimation |
|---|---|---|---|
| A | 1.49 | 0.92–2.09 | CA (100%) |
| B | 2.12 | 1.41–2.86 | CA (100%) |
| C | 3.06 | 2.15–4.02 | CA (100%) |
| D | 2.27 | 1.22–3.35 | CA (100%) |
| E | 2.72 | 1.80–3.69 | CA (100%) |
| F | 4.79 | 3.29–6.47 | CA (100%) |
| G | 1.41 | 0.59–2.47 | CA (100%) |
| H | 6.01 | 4.11–8.10 | CA-SA (95.8%), CA (4.2%) |
| I | 2.75 | 1.30–4.41 | CA-SA (100%) |
| J | 10.6 | 7.02–14.64 | SA (69.7%), CA-SA (30.3%) |
| Q | 15.55 | 10.68–21.23 | CA-SA (14.5%), SA (85.5%) |
| R | 21.12 | 14.21–28.95 | CA-SA (11.3%), SA (88.7%) |
4. Discussion
4.1. Phylogenetic Systematics and Biogeography
4.2. Geographic Color Pattern Variation
4.3. Taxonomic and Conservation Implications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stuart, S.; Chanson, J.S.; Cox, N.A.; Young, B.E.; Rodrigues, A.S.L.; Fishman, D.L.; Waller, R.W. Status and trends of amphibian declines and extinctions worldwide. Science 2004, 306, 1783–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, M.; Hilton-Taylor, C.; Angulo, A.; Böhm, M.; Brooks, T.M.; Butchart, S.H.M.; Carpenter, K.E.; Chanson, J.; Collen, B.; Cox, N.A.; et al. The impact of conservation on the status of the world’s vertebrates. Science 2010, 330, 1503–1509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-del-Pliego, P.; Freckleton, R.P.; Edwards, D.P.; Koo, M.S.; Scheffers, B.R.; Pyron, R.A.; Jetz, W. Phylogenetic and trait-based prediction of extinction risk for data-deficient amphibians. Curr. Biol. 2019, 29, 1557–1563. [Google Scholar] [CrossRef]
- Hayes, T.B.; Falso, P.; Gallipeau, S.; Stice, M. The cause of global amphibian declines: A developmental endocrinologist’s perspective. J. Exp. Biol. 2010, 213, 921–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skerratt, L.F.; Berger, L.; Speare, R.; Cashins, S.; McDonald, K.R.; Phillott, A.D.; Hines, H.B.; Kenyon, N. Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. EcoHealth 2007, 4, 125–134. [Google Scholar] [CrossRef]
- Scheele, B.C.; Pasmans, F.; Skerratt, L.F.; Berger, L.; Martel, A.; Beukema, W.; Acevedo, A.A.; Burrowes, P.A.; Carvalho, T.; Catenazzi, A.; et al. Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity. Science 2019, 363, 1459–1463. [Google Scholar] [CrossRef]
- Ron, S.R.; Duellman, W.E.; Coloma, L.A.; Bustamante, M.R. Population decline of the Jambato toad Atelopus ignescens (Anura: Bufonidae) in the Andes of Ecuador. J. Herpetol. 2003, 37, 116–126. [Google Scholar] [CrossRef]
- Coloma, L.A.; Duellman, W.E.; Almendáriz C., A.; Ron, S.R.; Terán-Valdez, A.; Guayasamin, J.M. Five new (extinct?) species of Atelopus (Anura: Bufonidae) from Andean Colombia, Ecuador, and Peru. Zootaxa 2010, 2574, 1–54. [Google Scholar] [CrossRef]
- Guayasamin, J.M.; Bonaccorso, E.; Duellman, W.E.; Coloma, L.A. Genetic differentiation in the nearly extinct harlequin frogs (Bufonidae: Atelopus), with emphasis on the Andean Atelopus ignescens and A. bomolochos species complexes. Zootaxa 2010, 2574, 55–68. [Google Scholar] [CrossRef]
- Lötters, S.; Meijden, A.; Coloma, L.A.; Boistel, R.; Cloetens, P.; Ernst, R.; Lehr, E.; Veith, M. Assessing the molecular phylogeny of a near extinct group of vertebrates: The Neotropical harlequin frogs (Bufonidae: Atelopus). Syst. Biodivers. 2011, 9, 45–57. [Google Scholar] [CrossRef]
- DiRenzo, G.V.; Langhammer, P.F.; Zamudio, K.R.; Lips, K.R. Fungal infection intensity and zoospore output of Atelopus zeteki, a potential acute chytrid supershedder. PLoS ONE 2014, 9, e93356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiRenzo, G.V.; Tunstall, T.S.; Ibáñez, R.; deVries, M.S.; Longo, A.V.; Zamudio, K.R.; Lips, K.R. External reinfection of a fungal pathogen does not contribute to pathogen growth. EcoHealth 2018, 15, 815–826. [Google Scholar] [CrossRef] [PubMed]
- La Marca, E.; Lötters, S.; Puschendorf, R.; Ibáñez, R.; Rueda-Almonacid, J.V.; Schulte, R.; Marty, C.; Castro, F.; Manzanilla-Puppo, J.; García-Pérez, J.E.; et al. Catastrophic population declines and extinctions in neotropical harlequin frogs (Bufonidae: Atelopus). Biotropica 2005, 37, 190–201. [Google Scholar] [CrossRef]
- Woodhams, D.C.; Alford, R.A.; Briggs, C.J.; Johnson, M.; Rollins-Smith, L.A. Life-history trade-offs influence disease in changing climates: Strategies of an amphibian pathogen. Ecology 2008, 89, 1627–1639. [Google Scholar] [CrossRef] [Green Version]
- Savage, J.M. The Amphibians and Reptiles of Costa Rica: A Herpetofauna between Two Continents, between Two Seas; University of Chicago Press: Chicago, IL, USA, 2002. [Google Scholar]
- Savage, J.M.; Bolaños, F. An enigmatic frog of the genus Atelopus (Family Bufonidae) from Parque Nacional Chirripó, Cordillera de Talamanca, Costa Rica. Rev. Biol. Trop. 2009, 57, 381–386. [Google Scholar] [CrossRef] [Green Version]
- Köhler, G. Amphibians of Central America; Herpeton Verlag: Offenbach, Germany, 2011. [Google Scholar]
- IUCN The IUCN Red List of Threatened Species. Available online: www.iucnredlist.org (accessed on 10 January 2019).
- Gagliardo, R.; Crump, P.; Griffith, E.; Mendelson, J.; Ross, H.; Zippel, K. The principles of rapid response for amphibian conservation, using the programmes in Panama as an example. Int. Zoo Yearb. 2008, 42, 125–135. [Google Scholar] [CrossRef]
- Zippel, K.; Johnson, K.; Gagliardo, R.; Gibson, R.; McFadden, M.; Browne, R.; Martinez, C.; Townsend, E. The Amphibian Ark: A global community for ex situ conservation of amphibians. Herpetol. Conserv. Biol. 2011, 6, 340–352. [Google Scholar]
- Estrada, A.; Gratwicke, B.; Benedetti, A.; DellaTogna, G.; Garrelle, G.; Griffith, E.; Ibáñez, R.; Ryan, S.; Miller, P.S. The Golden Frogs of Panama (Atelopus zeteki, A. varius): A Conservation Planning Workshop; IUSN/SSC Conservation Breeding Specialist Group: Apple Valley, MN, USA, 2014. [Google Scholar]
- Cikanek, S.J.; Nockold, S.; Brown, J.L.; Carpenter, J.W.; Estrada, A.; Guerrel, J.; Hope, K.; Ibáñez, R.; Putman, S.B.; Gratwicke, B. Evaluating group housing strategies for the ex-situ conservation of harlequin frogs (Atelopus spp.) using behavioral and physiological indicators. PLoS ONE 2014, 9, e90218. [Google Scholar] [CrossRef]
- Lewis, C.H.R.; Richards-Zawacki, C.L.; Ibáñez, R.; Luedtke, J.; Voyles, J.; Houser, P.; Gratwicke, B. Conserving Panamanian harlequin frogs by integrating captive-breeding and research programs. Biol. Conserv. 2019, 236, 180–187. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Luikart, G.H.; Aitken, S.N. Conservation and the Genetics of Populations, 2nd ed.; John Wiley & Sons, Ltd.: West Sussex, UK, 2012. [Google Scholar]
- Daugherty, C.H.; Cree, A.; Hay, J.M.; Thompson, M.B. Neglected taxonomy and continuing extinctions of tuatara (Sphenodon). Nature 1990, 347, 177–179. [Google Scholar] [CrossRef]
- Frankham, R.; Briscoe, D.A.; Ballou, J.D. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar]
- Noonan, B.P.; Gaucher, P. Phylogeography and demography of Guianan harlequin toads (Atelopus): Diversification within a refuge. Mol. Ecol. 2005, 14, 3017–3031. [Google Scholar] [CrossRef] [PubMed]
- Richards, C.L.; Knowles, L.L. Tests of phenotypic and genetic concordance and their application to the conservation of Panamanian golden frogs (Anura, Bufonidae). Mol. Ecol. 2007, 16, 3119–3133. [Google Scholar] [CrossRef] [PubMed]
- Lötters, S.; van der Meijden, A.; Rödder, D.; Köster, T.E.; Kraus, T.; La Marca, E.; Haddad, C.F.B.; Veith, M. Reinforcing and expanding the predictions of the disturbance vicariance hypothesis in Amazonian harlequin frogs: A molecular phylogenetic and climate envelope modelling approach. Biodivers. Conserv. 2010, 19, 2125–2146. [Google Scholar] [CrossRef] [Green Version]
- Stehli, F.G.; Webb, S.D. The Great American Biotic Interchange; Plenum Press: New York, NY, USA, 1985. [Google Scholar]
- Cody, S.; Richardson, J.E.; Rull, V.; Ellis, C.; Pennington, R.T. The Great American Biotic Interchange revisited. Ecography 2010, 33, 326–332. [Google Scholar] [CrossRef]
- Keigwin, J.D., Jr. Pliocene closing of the Isthmus of Panama, based on biostratigraphic evidence from nearby Pacific Ocean and Caribbean Sea cores. Geology 1978, 6, 630–634. [Google Scholar] [CrossRef]
- Coates, A.G.; Obando, J.A. The geologic evolution of the Central American Isthmus. In Evolution and Environment in Tropical America; Jackson, J.B.C., Budd, A.F., Coates, A.G., Eds.; University of Chicago Press: Chicago, IL, USA, 1996; pp. 21–56. [Google Scholar]
- Montes, C.; Cardona, A.; Jaramillo, C.; Pardo, A.; Silva, J.C.; Valencia, V.; Ayala, C.; Pérez-Angel, L.C.; Ramírez, V.; Niño, H. Middle Miocene closure of the Central American seaway. Science 2015, 348, 226–229. [Google Scholar] [CrossRef] [Green Version]
- Bacon, C.D.; Silvestro, D.; Jaramillo, C.; Smith, B.T.; Chakrabarty, P.; Antonelli, A. Biological evidence supports an early and complex emergence of the Isthmus of Panama. Proc. Natl. Acad. Sci. USA 2015, 112, 6110–6115. [Google Scholar] [CrossRef] [Green Version]
- O’Dea, A.; Lessios, H.A.; Coates, A.G.; Eytan, R.I.; Restrepo-Moreno, S.A.; Cione, A.L.; Collins, L.; de Queiroz, K.; Farris, D.W.; Norris, R.D.; et al. Formation of the Isthmus of Panama. Sci. Adv. 2016, 2, e1600883. [Google Scholar] [CrossRef] [Green Version]
- Daza, J.M.; Smith, E.N.; Páez, V.P.; Parkinson, C.L. Complex evolution in the Neotropics: The origin and diversification of the widespread genus Leptodeira (Serpentes: Colubridae). Mol. Phylogenet. Evol. 2009, 53, 653–667. [Google Scholar] [CrossRef]
- Pinto-Sánchez, N.R.; Ibáñez, R.; Madriñán, S.; Sanjur, O.I.; Bermingham, E.; Crawford, A.J. The Great American Biotic Interchange in frogs: Multiple and early colonization of Central America by the South American genus Pristimantis (Anura: Craugastoridae). Mol. Phylogenet. Evol. 2012, 62, 954–972. [Google Scholar] [CrossRef]
- Elmer, K.R.; Bonett, R.M.; Wake, D.B.; Lougheed, S.C. Early Miocene origin and cryptic diversification of South American salamanders. BMC Evol. Biol. 2013, 13, 59. [Google Scholar] [CrossRef] [Green Version]
- Palumbi, S.R. Nucleic acids II: The polymerase chain reaction. In Molecular Systematics; Hillis, D.M., Moritz, C., Mable, B.K., Eds.; Sinauer Associates: Sunderland, MA, USA, 1996; pp. 205–247. [Google Scholar]
- Kessing, B.; Croom, H.; Martin, A.; McIntosh, C.; McMillan, W.O.; Palumbi, S. The Simple Fool’s Guide to PCR, Version 1.0; University of Hawaii: Honolulu, HI, USA, 1989. [Google Scholar]
- Crawford, A.J.; Bermingham, E.; Polanía, C. The role of tropical dry forest as a long-term barrier to dispersal: A comparative phylogeographical analysis of dry forest tolerant and intolerant frogs. Mol. Ecol. 2007, 16, 4789–4807. [Google Scholar] [CrossRef] [PubMed]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.10. Available online: http://mesquiteproject.org (accessed on 1 September 2016).
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods); Version 4; Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Lanfear, R.; Calcott, B.; Ho, S.Y.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [Green Version]
- Zwickl, D.J. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets under the Maximum Likelihood Criterion. Ph.D. Thesis, The University of Texas at Austin, Austin, TX, USA, 2006. [Google Scholar]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouckaert, R.; Alvarado-Mora, M.V.; Pinho, J.R. Evolutionary rates and HBV: Issues of rate estimation with Bayesian molecular methods. Antivir. Ther. 2013, 18, 497–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gernhard, T. The conditioned reconstructed process. J. Theor. Biol. 2008, 253, 769–778. [Google Scholar] [CrossRef] [Green Version]
- Macey, J.R.; Schulte, J.A., II; Larson, A.; Fang, Z.; Wang, Y.; Tuniyev, B.S.; Papenfuss, T.J. Phylogenetic relationships of toads in the Bufo bufo species group from the eastern escarpment of the Tibetan Plateau: A case of vicariance and dispersal. Mol. Phylogenet. Evol. 1998, 9, 80–87. [Google Scholar] [CrossRef] [Green Version]
- Crawford, A.J. Relative rates of nucleotide substitution in frogs. J. Mol. Evol. 2003, 57, 636–641. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Matzke, N.J. BioGeoBEARS’: Biogeography with Bayesian (and Likelihood) Evolutionary Analysis in R Scripts. Available online: http://CRAN.R-project.org/package=BioGeoBEARS (accessed on 21 February 2018).
- Matzke, N.J. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst. Biol. 2014, 63, 951–970. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Fouquet, A.; Gilles, A.; Vences, M.; Marty, C.; Blanc, M.; Gemmel, N.J. Underestimation of species richness in Neotropical frogs revealed by mtDNA analyses. PLoS ONE 2007, 2, e1109. [Google Scholar] [CrossRef] [PubMed]
- Vences, M.; Thomas, M.; Bonett, R.M.; Vieites, D.R. Deciphering amphibian diversity through DNA barcoding: Chances and challenges. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1859–1868. [Google Scholar] [CrossRef] [Green Version]
- Flechas, S.V.; Sarmiento, C.; Cárdenas, M.E.; Medina, E.M.; Restrepo, S.; Amézquita, A. Surviving chytridiomycosis: Differential anti-Batrachochytrium dendrobatidis activity in bacterial isolates from three lowland species of Atelopus. PLoS ONE 2012, 7, e44832. [Google Scholar] [CrossRef] [Green Version]
- Savage, J.M. The harlequin frogs, genus Atelopus, of Costa Rica and western Panama. Herpetologica 1972, 28, 77–94. [Google Scholar]
- Lötters, S.; Böhme, W.; Günther, R. Notes on the type material of the neotropical harlequin frogs Atelopus varius (Lichtenstein & Martens, 1856) and Atelopus cruciger (Lichtenstein & Martens, 1856) deposited in the Museum für Naturkunde of Berlin (Anura, Bufonidae). Mitteilungen Aus Dem Mus. Für Naturkunde Berl. Zool. Reihe 1998, 74, 173–184. [Google Scholar]
- Rodríguez, C.; Rollins-Smith, L.; Ibáñez, R.; Durant-Archibold, A.A.; Gutiérrez, M. Toxins and pharmacologically active compounds from species of the family Bufonidae (Amphibia, Anura). J. Ethnopharmacol. 2017, 198, 235–254. [Google Scholar] [CrossRef] [Green Version]
- Wells, K.D. The Ecology and Behavior of Amphibians; University of Chicago Press: Chicago, IL, USA, 2007. [Google Scholar]
- Ree, R.H.; Smith, S.A. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 2008, 57, 4–14. [Google Scholar] [CrossRef] [Green Version]
- Castroviejo-Fisher, S.; Guayasamin, J.M.; Gonzalez-Voyer, A.; Vila, C. Neotropical diversification seen through glassfrogs. J. Biogeogr. 2014, 4, 66–80. [Google Scholar] [CrossRef] [Green Version]
- Winston, M.E.; Kronaure, D.J.C.; Moreau, C.S. Early and dynamic colonization of Central America drives speciation in Neotropical army ants. Mol. Ecol. 2017, 26, 859–870. [Google Scholar] [CrossRef]
- Warburg, M.R. Ecophysiology of Amphibians Inhabiting Xeric Environments; Springer: Berlin, Germany, 1997. [Google Scholar]
- Cruz-Piedrahita, C.; Navas, C.A.; Crawford, A.J. Life on the edge: A comparative study of ecophysiological adaptations of frogs to tropical semiarid environments. Physiol. Biochem. Zool. 2018, 91, 740–756. [Google Scholar] [CrossRef] [Green Version]
- Bacon, C.D.; Molnar, P.; Antonelli, A.; Crawford, A.J.; Montes, C.; Vallejo-Pareja, M.C. Quaternary glaciation and the Great American Biotic Interchange. Geology 2016, 44, 375–378. [Google Scholar] [CrossRef] [Green Version]
- Zippel, K.; Ibáñez, D.R.; Lindquist, E.D.; Richards, C.L.; Jaramillo, A.C.A.; Griffith, E.J. Implicaciones en la conservación de las ranas dorados de Panamá, asociadas con su revisión taxonómica. Herpetotropicos 2006, 3, 29–39. [Google Scholar]
- Lindquist, E.D.; Hetherington, T.E. Tadpoles and juveniles of the Panamanian golden frog, Atelopus zeteki (Bufonidae), with information on development of coloration and patterning. Herpetologica 1998, 54, 370–376. [Google Scholar]
- Ibáñez, D.R.; Jaramillo, C.A.; Solís, F.A. Una especie nueva de Atelopus (Amphibia: Bufonidae) de Panamá. Caribb. J. Sci. 1995, 31, 57–64. [Google Scholar]
- Dunn, E.R. New frogs from Panama and Costa Rica. Occas. Pap. Boston Soc. Nat. Hist. 1931, 5, 385–401. [Google Scholar]
- Cochran, D.M.; Goin, C.J. Frogs of Colombia. U. S. Natl. Mus. Bull. 1970, 288, 1–655. [Google Scholar] [CrossRef]
- Lynch, J.D.; Suárez-Mayorga, A. Catálogo de anfibios en el Chocó Biogeográfico. In Colombia Diversidad Biótica IV, El Chocó Biogeográfico/Costa Pacífica; Rangel, O., Ed.; Universidad Nacional de Colombia: Bogotá, Colombia, 2004; Volume I, pp. 654–668. [Google Scholar]
- Pounds, J.A. Amphibians and reptiles. In Monteverde: Ecology and Conservation of a Tropical Cloud Forest; Nadkarni, N.M., Wheelwright, N.T., Eds.; Oxford University Press: New York, NY, USA, 2000; pp. 149–177. [Google Scholar]
- Crawford, A.J.; Cruz, C.; Griffith, E.; Ross, H.; Ibáñez, R.; Lips, K.R.; Driskell, A.C.; Bermingham, E.; Crump, P. DNA barcoding applied to ex situ tropical amphibian conservation programme reveals cryptic diversity in captive populations. Mol. Ecol. Resour. 2013, 13, 1005–1018. [Google Scholar] [CrossRef]




| Gene | Length of Sequence (bp) | Invariant Sites | Singletons | Parsimony-Informative Sites |
|---|---|---|---|---|
| cyt b | 714 | 537 | 35 | 142 |
| COI | 639 | 493 | 10 | 136 |
| Taxon | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A. sp. “Puerto Obaldía-Capurganá” | 0–0.79 | 7.94–8.59 | 7.45–7.93 | 9.17–10.60 | 10.9–11.4 | 8.58–8.91 | 9.23–10.80 | 9.57–11.80 | 10.1–10.80 |
| 0–0.86 | ||||||||||
| 2 | A. certus | 0–0.63 | 2.91–3.16 | 7.58–9.60 | 9.78–9.96 | 7.77–7.80 | 7.95–9.11 | 7.31–8.26 | 8.12–8.45 | |
| 4.55–5.40 | 0–0.28 | |||||||||
| 3 | A. glyphus | 0 | 7.58–9.11 | 9.93–10.10 | 7.62–7.79 | 8.28–9.10 | 7.93–8.90 | 8.44–8.77 | ||
| 4.89–5.22 | 1.43–2.07 | 0–0.42 | ||||||||
| 4 | A. senex | 0.16–1.33 | 7.00–8.68 | 4.06–5.19 | 5.90–8.42 | 5.12–8.44 | 6.86–8.18 | |||
| 6.09–7.14 | 6.78–8.22 | 6.59–7.32 | 0–1.92 | |||||||
| 5 | A. sp. “Monteverde” (MVZ 164818) | 0 | 5.27 | 6.36–7.00 | 6.67–7.46 | 7.00–7.32 | ||||
| 6.80–7.13 | 6.61–7.31 | 6.43 | 4.62–4.91 | 0 | ||||||
| 6 | A. chiriquiensis | 0–0.16 | 3.46–4.37 | 4.20–5.58 | 5.13–5.75 | |||||
| 5.76–6.27 | 6.80–7.33 | 6.62–6.80 | 4.91–5.53 | 4.73–4.90 | 0–0.28 | |||||
| 7 | A. limosus | 0.31–1.26 | 3.75–5.27 | 4.36–5.91 | ||||||
| 5.92–6.79 | 6.09–7.49 | 5.92–6.62 | 4.72–5.93 | 5.93–6.28 | 5.08–5.43 | 0.28–1.42 | ||||
| 8 | A. varius | 0–2.08 | 2.86–4.66 | |||||||
| 6.12–7.00 | 6.28–8.25 | 5.92–6.98 | 4.81–5.96 | 5.56–6.29 | 4.24–4.92 | 3.89–4.74 | 0–2.44 | |||
| 9 | A. zeteki | 0–0.79 | ||||||||
| 6.44–7.13 | 6.63–7.69 | 6.09–6.44 | 5.15–6.45 | 5.92–6.63 | 4.73–5.24 | 3.71–4.89 | 2.72–3.72 | 0–0.28 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez, J.P.; Jaramillo, C.A.; Lindquist, E.D.; Crawford, A.J.; Ibáñez, R. Recent and Rapid Radiation of the Highly Endangered Harlequin Frogs (Atelopus) into Central America Inferred from Mitochondrial DNA Sequences. Diversity 2020, 12, 360. https://doi.org/10.3390/d12090360
Ramírez JP, Jaramillo CA, Lindquist ED, Crawford AJ, Ibáñez R. Recent and Rapid Radiation of the Highly Endangered Harlequin Frogs (Atelopus) into Central America Inferred from Mitochondrial DNA Sequences. Diversity. 2020; 12(9):360. https://doi.org/10.3390/d12090360
Chicago/Turabian StyleRamírez, Juan P., César A. Jaramillo, Erik D. Lindquist, Andrew J. Crawford, and Roberto Ibáñez. 2020. "Recent and Rapid Radiation of the Highly Endangered Harlequin Frogs (Atelopus) into Central America Inferred from Mitochondrial DNA Sequences" Diversity 12, no. 9: 360. https://doi.org/10.3390/d12090360