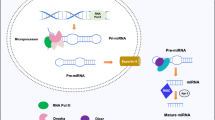
Abstract
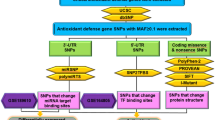
The process of genetically programmed cell death, or apoptosis, plays a crucialrolein cellular homeostasis and gene expression. Disruption of apoptosis may lead to aberrant immune responses, cancer, and neurodegenerative diseases. Single nucleotide polymorphisms (SNPs) present in various microRNA (miRNA) genes and targets being an alteration of miRNA activity resulting in human diseases. Evidence reported that SNPs increase/decrease the effectiveness of the interaction between miRNAs and their target genes associated with diseases. The primary purpose of this study is not only to identify miRSNPs on the CASP7 gene (caspase-7) and SNPs in miRNA genes targeting 3′UTR but also to evaluate the effect of thesegene variations in apoptosis and their associated diseases. We detected 120 miRNAs binding sites and 27 different SNPs in binding sites of miRNA in 3′UTR of the CASP7 gene by ten different online softwares. Interestingly, miR-371b-5p’s binding site on CASP7 has an SNP (rs576198588, G/T) on CASP7 3′UTR, and its genomic sequence has an SNP (rs751339395, G/T) at the same nucleotide with rs576198588. Similarly, two other SNPs (rs774879764, C/G rs750389063, C/T) were identified at the first position binding site of miR-371b-5p. Here, miRSNP (rs576198588) at CASP7 3′UTR and SNP (rs751339395) at miR-371b-5p genomic sequence cross-matches at the same site of binding region. Besides, miR-371b-5p targets many apoptosis-related genes (HIP1, TRIAP1, GSKIP, NIN, DAP, CAAP1, XIAP, TMBIM1, TMBIM4, TNFRSF10A, RAD21, AKT1, BAG1, BAG4) even though it had no apoptosis correlated interaction demonstrated formerly. It assures that CASP7 could have a significant consequence on apoptosis through different pathways. Henceforth, this study was representing and signifying an influential connotation among miR-371b-5p and apoptosis via computational exploration and recommended to have better insight.


Similar content being viewed by others
Data availability
All relevant data are within the paper and its supporting information files.
References
Lowe, S. W., & Lin, A. W. (2000). Apoptosis in cancer. Carcinogenesis, 21, 485–495.
Khailany, R. A., Safdar, & Ozaslan, M. (2019). Molecular investigation of KRAS gene in breast cancer patients. Journal of Biological Sciences, 19, 323–327.
Coutinho-Camillo, C. M., & Soares, F. A. (2015). CASP7 (caspase 7, apoptosis-related cysteine peptidase). Atlas of Genetics and Cytogenetics in Oncology and Haematology, 19(3), 160–163. https://doi.org/10.4267/2042/56404.
Nabholz, B., Ellegren, H., & Wolf, J. B. (2012). High levels of gene expression explain the strong evolutionary constraint of mitochondrial protein-coding genes. Molecular Biology and Evolution, 30, 272–284.
Bartke, T., Pohl, C., Pyrowolakis, G., & Jentsch, S. (2004). Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Molecular Cell, 14, 801–811.
Denault, J.-B., Békés, M., Scott, F. L., Sexton, K. M., Bogyo, M., & Salvesen, G. S. (2006). Engineered hybrid dimers: tracking the activation pathway of caspase-7. Molecular Cell, 23, 523–533.
Tenev, T., Zachariou, A., Wilson, R., Ditzel, M., & Meier, P. (2005). IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nature Cell Biology, 7, 70–77.
Ruiz‐Vela, A., de Buitrago, G. G., & Martínez, C. (1999). Implication of calpain in caspase activation during B cell clonal deletion. The EMBO Journal, 18, 4988–4998.
Thiantanawat, A., Long, B. J., & Brodie, A. M. (2003). Signaling pathways of apoptosis activated by aromatase inhibitors and antiestrogens. Cancer Research, 63, 8037–8050.
Enari, M., Sakahira, H., Yokoyama, H., Okawa, K., Iwamatsu, A., & Nagata, S. (1998). A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature, 391, 43–50.
Li, X., Marani, M., Yu, J., Nan, B., Roth, J. A., & Kagawa, S., et al. (2001). Adenovirus-mediated Bax overexpression for the induction of therapeutic apoptosis in prostate cancer. Cancer Research, 61, 186–191.
BrÁs, A., Ruiz-vela, A., De Buitrago, G. G., & MartÍnez-a, C. (1999). Caspase activation by BCR cross-linking in immature B cells: differential effects on growth arrest and apoptosis. The FASEB Journal, 13, 931–944.
Chandler, J. M., Cohen, G. M., & MacFarlane, M. (1998). Different subcellular distribution of caspase-3 and caspase-7 following Fas-induced apoptosis in mouse liver. Journal of Biological Chemistry, 273, 10815–10818.
Chandler, J. M., Alnemri, E. S., Cohen, G. M., & MacFarlane, M. (1997). Activation of CPP32 and Mch3α in wild-type p53-induced apoptosis. Biochemical Journal, 322, 19–23.
Kimura, F., Suzu, S., Nakamura, Y., Nakata, Y., Yamada, M., & Kuwada, N., et al. (2003). Cloning and characterization of a novel RING-B-box-coiled-coil protein with apoptotic function. Journal of Biological Chemistry, 278, 25046–25054.
Korfali, N., Ruchaud, S., Loegering, D., Bernard, D., Dingwall, C., & Kaufmann, S. H., et al. (2004). Caspase-7 gene disruption reveals an involvement of the enzyme during the early stages of apoptosis. Journal of Biological Chemistry, 279, 1030–1039.
Arshad, M., Ozaslan, M., Ali, H. K., Safdar, M., Junejo, Y., & Babar, M. E. (2019). Molecular Investigation of Gold Nanoparticles Toxicity in Mice Model and p53 Activation. Journal of Biological Sciences, 19, 391–395.
Nakajima, G., Hayashi, K., Xi, Y., Kudo, K., Uchida, K., & Takasaki, K., et al. (2006). Non-coding microRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer Genomics-Proteomics, 3, 317–324.
Ergun, S., & Oztuzcu, S. (2014). Computational analysis of 3′ UTR Region of CASP3 with respect to miRSNPs and SNPs in targetting miRNAs. Computational Biology and Chemistry, 53, 235–241.
Paraskevopoulou, M. D., Georgakilas, G., Kostoulas, N., Vlachos, I. S., Vergoulis, T., & Reczko, M., et al. (2013). DIANA-microT web server v5. 0: service integration into miRNA functional analysis workflows. Nucleic Acids Research, 41, W169–W173.
Vlachos, I. S., Kostoulas, N., Vergoulis, T., Georgakilas, G., Reczko, M., & Maragkakis, M., et al. (2012). DIANA miRPath v. 2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Research, 40, W498–W504.
Hsu, S.-D., Tseng, Y.-T., Shrestha, S., Lin, Y.-L., Khaleel, A., & Chou, C.-H., et al. (2014). miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Research, 42, D78–D85.
Betel, D., Koppal, A., Agius, P., Sander, C., & Leslie, C. (2010). Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biology, 11, R90.
Bommer, G. T., Gerin, I., Feng, Y., Kaczorowski, A. J., Kuick, R., & Love, R. E., et al. (2007). p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Current Biology, 17, 1298–1307.
Wong, N., & Wang, X. (2014). miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Research, 43, D146–D152.
Wang, R., Wang, S. T., Wang, Y. D., Wu, G., Du, Y., & Qian, M. Q., et al. (2016). Stress-responsive heme oxygenase-1 isoenzyme participates in Toll-like receptor 4-induced inflammation during brain ischemia. Neuroreport, 27, 445–454.
Xiao, F., Zuo, Z., Cai, G., Kang, S., Gao, X., & Li, T. (2008). miRecords: an integrated resource for microRNA–target interactions. Nucleic Acids Research, 37, D105–D110.
Jeggari, A., Marks, D. S., & Larsson, E. (2012). miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics, 28, 2062–2063.
Huang, Y., Zou, Q., Song, H., Song, F., Wang, L., & Zhang, G., et al. (2010). A study of miRNAs targets prediction and experimental validation. Protein & Cell, 1, 979–986.
Gosline, S. J., Gurtan, A. M., JnBaptiste, C. K., Bosson, A., Milani, P., & Dalin, S., et al. (2016). Elucidating MicroRNA regulatory networks using transcriptional, post-transcriptional, and histone modification measurements. Cell Reports, 14, 310–319.
Cui, R., Kim, T., Fassan, M., Meng, W., Sun, H.-L., & Jeon, Y.-J., et al. (2015). MicroRNA-224 is implicated in lung cancer pathogenesis through targeting caspase-3 and caspase-7. Oncotarget, 6, 21802.
Lee, Y. S., Nakahara, K., Pham, J. W., Kim, K., He, Z., & Sontheimer, E. J., et al. (2004). Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell, 117, 69–81.
Cai, X., Hagedorn, C. H., & Cullen, B. R. (2004). Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA, 10, 1957–1966.
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297.
Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., & Noch, E., et al. (2002). Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences, 99, 15524–15529.
Baek, D., Villén, J., Shin, C., Camargo, F. D., Gygi, S. P., & Bartel, D. P. (2008). The impact of microRNAs on protein output. Nature, 455, 64–71.
Ruan, W., Xu, J.-m, Li, S.-b, Yuan, L.-q, & Dai, R.-p (2011). Effects of down-regulation of microRNA-23a on TNF-α-induced endothelial cell apoptosis through caspase-dependent pathways. Cardiovascular Research, 93, 623–632.
Scott, G. K., Goga, A., Bhaumik, D., Berger, C. E., Sullivan, C. S., & Benz, C. C. (2007). Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. Journal of Biological Chemistry, 282, 1479–1486.
Chen, G., Umelo, I. A., Lv, S., Teugels, E., Fostier, K., & Kronenberger, P., et al. (2013). miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PloS ONE, 8, e60317.
Tang, Y., Zhang, Y.-c., Chen, Y., Xiang, Y., Shen, C.-x., & Li, Y.-g. (2015). The role of miR-19b in the inhibition of endothelial cell apoptosis and its relationship with coronary artery disease. Scientific Reports, 5, 15132. https://doi.org/10.1038/srep15132.
Ohira, T., Naohiro, S., Nakayama, Y., Osaki, M., Okada, F., & Oshi-mura, M., et al. (2015). miR-19b regulates hTERT mRNA expression through targeting PITX1 mRNA in melanoma cells. Scientific Reports, 5, 8201. https://doi.org/10.1038/srep08201.
Huh, J., Kim, T., Kim, K., Song, J., Jung, Y., & Jeong, J., et al. (2013). Dysregulation of miR-106a and miR-591 confers paclitaxel resistance to ovarian cancer. British Journal of Cancer, 109, 452–461.
Mi, Y., Wang, L., Zong, L., Pei, M., & Lu, Q., et al. (2014). Genetic variants in microRNA target sites of 37 selected cancer-related genes and the risk of cervical cancer. PLoS ONE, 9(1), e86061. https://doi.org/10.1371/journal.pone.0086061.
Mi, Y., Zhang, D., Jiang, W., Weng, J., Zhou, C., & Huang, K., et al. (2017). miR-181a-5p promotes the progression of gastric cancer via RASSF6-mediated MAPK signalling activation. Cancer Letters, 389, 11–22.
Xu, H. L., Xu, W. H., Cai, Q., Feng, M., Long, J., & Zheng, W., et al. (2009). Polymorphisms and haplotypes in the caspase-3, caspase-7, and caspase-8 genes and risk for endometrial cancer: a population-based, case-control study in a Chinese population. Cancer Epidemiol Biomarkers Prev, 18, 2114–2122.
Shi, T.-Y., et al. (2015). CASP7 variants modify susceptibility to cervical cancer in Chinese women. Scientific Reports, 5, 9225. https://doi.org/10.1038/srep09225.
Lee, W. K., Kim, J. S., Kang, H. G., Cha, S. I., Kim, D. S., & Hyun, D. S., et al. (2009). Polymorphisms in the Caspase7 gene and the risk of lung cancer. Lung Cancer, 65, 19–24.
Jiang, W., Bi, N., & Zhang, W. J. (2016). MicroRNA-related polymorphisms in apoptosis pathway genes are predictive of clinical outcome in patients with limited disease small cell lung cancer. Oncotarget, 7(16), 22632–22638.
Li, S., Yao, W., Pan, Q., Tang, X., Zhao, S., Wang, W., Zhu, Z., Gao, J., Sheng, Y., & Zhou, F., et al. (2015). Association analysis revealed one susceptibility locus for vitiligo with immune-related diseases in the Chinese Han population. Immunogenetics, 67(7), 347–354. https://doi.org/10.1007/s00251-015-0843-4.
Fiorino, S., Bacchi-Reggiani, M. L., Visani, M., Acquaviva, G., Fornelli, A., Masetti, M., Tura, A., Grizzi, F., Zanello, M., Mastrangelo, L., Lombardi, R., Di Tommaso, L., & Bondi, A., et al. (2016). MicroRNAs as possible biomarkers for diagnosis and prognosis of hepatitis B- and C-related-hepatocellular-carcinoma. World J Gastroenterol, 22, 3907–3936.
Daly, J. M., Jannot, C. B., Beerli, R. R., Graus-Porta, D., Maurer, F. G., & Hynes, N. E. (1997). Neu differentiation factor induces ErbB2 down-regulation and apoptosis of ErbB2-overexpressing breast tumor cells. Cancer Research, 57, 3804–3811.
Iorio, M. V., Casalini, P., Piovan, C., Di Leva, G., Merlo, A., & Triulzi, T., et al. (2009). microRNA-205 regulates HER3 in human breast cancer. Cancer Research, 69, 2195–2200.
Soares, R., Meireles, M., Rocha, A., Pirraco, A., Obiol, D., & Alonso, E., et al. (2011). Maitake (D fraction) mushroom extract induces apoptosis in breast cancer cells by BAK-1 gene activation. Journal of Medicinal Food, 14, 563–572.
Beurel, E., Kornprobst, M., Blivet-Van Eggelpoël, M.-J., Ruiz-Ruiz, C., Cadoret, A., & Capeau, J., et al. (2004). GSK-3β inhibition by lithium confers resistance to chemotherapy-induced apoptosis through the repression of CD95 (Fas/APO-1) expression. Experimental Cell Research, 300, 354–364.
Bouker, K. B., Skaar, T. C., Riggins, R. B., Harburger, D. S., Fernandez, D. R., & Zwart, A., et al. (2005). Interferon regulatory factor-1 (IRF-1) exhibits tumor suppressor activities in breast cancer associated with caspase activation and induction of apoptosis. Carcinogenesis, 26, 1527–1535.
Kannan, K., Kaminski, N., Rechavi, G., Jakob-Hirsch, J., Amariglio, N., & Givol, D. (2001). DNA microarray analysis of genes involved in p53 mediated apoptosis: activation of Apaf-1. Oncogene, 20, 3449.
杨隽, 司书毅, 张月琴. Smad 在 TGF-β 超家族信号通路中的调控作用. 中国生物工程杂志 2003;23,9–12.
Acknowledgements
All the authors of the paper thank and acknowledge their respective Universities and Institutes.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Safdar, M., Zaheer, S., Khailany, R.A. et al. The Relevance of SNPs at 3′UTR Region of CASP7 and miR-371b-5p Associated Diseases: A Computational Analysis. Cell Biochem Biophys 78, 541–557 (2020). https://doi.org/10.1007/s12013-020-00941-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-020-00941-2