Antibody Modified Gold Electrode as an Impedimetric Biosensor for the Detection of Streptococcus pyogenes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
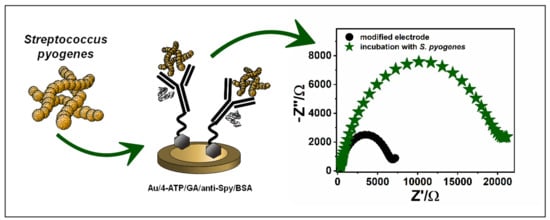
2.2. Preparation of the Biosensor
3. Results
3.1. Anti-Spy Immobilization on the Surface of the Electrodes
3.2. Electrochemical Detection of Streptococcus Pyogenes
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Sanyahumbi, A.S.; Colquhoun, S.; Wyber, R.; Carapetis, J.R. Global Disease Burden of Group A Streptococcus. In Basic Biology to Clinical Manifestations; Ferretti, J.J., Stevens, D.L., Fischetti, V.A., Eds.; University of Oklahoma Health Sciences Center: Oklahoma City, OK, USA, 2016; pp. 661–704. [Google Scholar]
- Cho, K.H.; Port, G.C.; Caparon, M. Genetics of Group A Streptococci. Microbiol. Spec. 2019, 7, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Carapetis, J.R.; Steer, A.C.; Mullholland, E.K.; Weber, M. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 2005, 5, 685–694. [Google Scholar] [CrossRef]
- Dajani, A.; Taubert, K.; Ferrieri, P.; Peter, G.; Shulman, S. Treatment of Acute Streptococcal Pharyngitis and Prevention of Rheumatic Fever: A Statement for Health Professionals. Pediatrics 1995, 96, 758–764. [Google Scholar] [PubMed]
- Leung, T.N.H.; Hon, K.L.; Leung, A.K.C. Group A Streptococcus disease in Hong Kong children: An overview. Hong Kong Med. J. 2018, 24, 593–601. [Google Scholar] [CrossRef]
- Bisno, A.L. Acute Pharyngitis. N. Eng. J. Med. 2001, 344, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Johansson, L.; Thulin, P.; Low, E.D.; Norrby-Teglund, A. Getting under the Skin: The Immunopathogenesis of Streptococcus pyogenes Deep Tissue Infections. Clin. Infect. Dis. 2010, 51, 58–65. [Google Scholar] [CrossRef] [Green Version]
- Kaushal, A.; Goyal, L.; Kumar, A. Recent advances in diagnosis of rheumatic heart disease. Biosci. Biotech. Res. Comm. 2011, 4, 1–9. [Google Scholar]
- Chandra, P.; Maurya, P.K.; Kumar, P.; Tripathi, P.; Srivastava, A.K. Diagnosis of Rheumatic Infections Caused by Group A Streptococcus Pyogenes: Future Investigation By Nanotechnology. Dig. J. Nanomater. Bios. 2009, 4, 645–650. [Google Scholar]
- Hytonen, J.; Haataja, S.; Gerlach, D.; Podbielski, A.; Finne, J. The SpeB virulence factor of Streptococcus pyogenes, a multifunctional secreted and cell surface molecule with strepadhesin, laminin-binding and cysteine protease activity. Mol. Microbiol. 2001, 39, 512–519. [Google Scholar] [CrossRef]
- Cunningham, M.W. Pathogenesis of Group A Streptococcal Infections. Clin. Microbiol. Rev. 2000, 13, 470–511. [Google Scholar] [CrossRef]
- Bisno, A.; Brito, M.; Collins, C. Molecular basis of group A streptococcal virulence. Lancet Infect. 2003, 3, 191–200. [Google Scholar] [CrossRef]
- Luo, R.; Sickler, J.; Vahidnia, F.; Lee, Y.C.; Frogner, B.; Thompson, M. Diagnosis and Management of Group a Streptococcal Pharyngitis in the United States. BMC Infect. Dis. 2019, 19, 193. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.J.; Barnett, T.C.; McArthur, J.D.; Cole, J.N.; Gillen, C.M.; Henningham, A.; Sriprakash, K.S.; Sanderson-Smith, M.L.; Nizet, V. Disease Manifestations and Pathogenic Mechanisms of Group A Streptococcus. Clin. Microbiol. Rev. 2014, 27, 264–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laabei, M.; Ermert, D. Catch Me if You Can: Streptococcus pyogenes Complement Evasion Strategies. J. Innate Immun. 2019, 11, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Jansen, J.; Rink, L. Streptococcus. In Molecular Detection of Human Bacterial Pathogens, 1st ed.; CRC Press: Florida, FL, USA, 2011; pp. 323–336. [Google Scholar]
- Spellerberg, B.; Brandt, C. Laboratory Diagnosis of Streptococcus pyogenes (group A streptococci). In Streptococcus pyogenes: Basic Biology to Clinical Manifestations; Ferretti, J.J., Stevens, D.L., Fischetti, V.A., Eds.; University of Oklahoma Health Sciences Center: Oklahoma City, OK, USA, 2016; pp. 931–946. [Google Scholar]
- Zhang, B.; Song, H.; Shen, Q.; Shi, Q.; Bai, S. Detection of Streptococcus pyogenes antibodies in acute idiopathic urticaria. Int. J. Clin. Exp. Med. 2017, 6, 10736–10741. [Google Scholar]
- Henson, A.M.; Carter, D.; Todd, K.; Shulman, S.T.; Zheng, X. Detection of Streptococcus pyogenes by Use of Illumigene Group A Streptococcus Assay. J. Clin. Microbiol. 2013, 51, 4207–4209. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhou, N.; Xu, B.; Hao, H.; Kang, L.; Zheng, Y.; Jiang, Y.; Jiang, H. Identification and Cluster Analysis of Streptococcus pyogenes by MALDI-TOF Mass Spectrometry. PLoS ONE 2012, 7, e47152. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; O’Leary, A.; Uhl, J.R.; Patel, R.; Shulman, S.T. Rapid Detection of Streptococcus pyogenes in Pleural Fluid Samples from Pediatric Patients with Empyema. J. Clin. Microbiol. 2012, 50, 2786–2787. [Google Scholar] [CrossRef] [Green Version]
- Uhl, J.R.; Adamson, S.C.; Vetter, E.A.; Schleck, C.D.; Harmsen, W.S.; Iverson, L.K.; Santrach, P.J.; Henry, N.K.; Cockerill, F.R. Comparison of LightCycler PCR, Rapid Antigen Immunoassay, and Culture for Detection of Group A Streptococci from Throat Swabs. J. Clin. Microbiol. 2003, 41, 242–249. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Tian, Y.; Chen, L.; Luo, R.; Sickler, J.; Liesenfeld, O.; Chen, S. Accurate Detection of Streptococcus pyogenes at the Point of Care Using the cobas Liat Strep A Nucleic Acid Test. Clin. Pediatr. 2017, 56, 1128–1134. [Google Scholar] [CrossRef]
- Slinger, R.; Goldfarb, D.; Rajakumar, D.; Moldovan, I.; Barrowman, N.; Tam, R.; Chan, F. Rapid PCR detection of group a streptococcus from flocked throat swabs: A retrospective clinical study. Ann. Clin. Microbiol. Antimicrob. 2011, 10, 33. [Google Scholar] [CrossRef] [Green Version]
- Borek, A.L.; Obszańska, K.; Hryniewicz, W.; Sitkiewicz, I. Detection of Streptococcus pyogenes virulence factors by multiplex PCR. Virulence 2012, 3, 529–533. [Google Scholar] [CrossRef] [Green Version]
- Uhl, J.R.; Patel, R. Fifteen-Minute Detection of Streptococcus pyogenes in Throat Swabs by Use of a Commercially Available Point-of-Care PCR Assay. J. Clin. Microbiol. 2016, 54, 815. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; He, F.; Lian, Y.; Yan, D.; Zhang, X. A new aptamer/SWNTs IDE-SPQC sensor for rapid and specific detection of Group A Streptococcus. Sens. Actuators B Chem. 2014, 198, 431–437. [Google Scholar] [CrossRef]
- Sotillo, A.; Pedrero, M.; de Pablos, M.; García, J.L.; García, E.; García, P.; Pingarrón, J.M.; Mingorance, J.; Campuzano, S. Clinical evaluation of a disposable amperometric magneto-genosensor for the detection and identification of Streptococcus pneumoniae. J. Microbiol. Methods 2014, 103, 25–28. [Google Scholar] [CrossRef]
- Singh, S.; Kaushal, A.; Khare, S.; Kumar, A. DNA chip based sensor for amperometric detection of infectious pathogens. Int. J. Biol. Macromol. 2017, 103, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Kaushal, A.; Khare, S.; Kumar, P.; Kumar, A. Gold–mercaptopropionic acid–polyethylenimine composite based DNA sensor for early detection of rheumatic heart disease. Analyst 2014, 139, 3600–3606. [Google Scholar] [CrossRef]
- Sheybani, R.; Shukla, A. Highly sensitive label-free dual sensor array for rapid detection of wound bacteria. Biosens. Bioelectron. 2017, 92, 425–433. [Google Scholar] [CrossRef] [Green Version]
- Lodes, M.J.; Suciu, D.; Wilmoth, J.L.; Ross, M.; Munro, S.; Dix, K.; Bernards, K.; Stöver, A.G.; Quintana, M.; Iihoshi, N.; et al. Identification of Upper Respiratory Tract Pathogens Using Electrochemical Detection on an Oligonucleotide Microarray. PLoS ONE 2007, 2, e924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaushal, A.; Singh, S.; Kumar, A.; Kumar, D. Nano-Au/cMWCNT Modified speB Gene Specific Amperometric Sensor for Rapidly Detecting Streptococcus pyogenes causing Rheumatic Heart Disease. Indian J. Microbiol. 2017, 57, 121–124. [Google Scholar] [CrossRef] [Green Version]
- Zheng, F.; Wang, P.; Du, Q.; Chen, Y.; Liu, N. Simultaneous and Ultrasensitive Detection of Foodborne Bacteria by Gold Nanoparticles-Amplified Microcantilever Array Biosensor. Front. Chem. 2019, 7, 232. [Google Scholar] [CrossRef] [Green Version]
- Kaushal, A.; Singh, S.; Kala, D.; Kumar, D.; Kumar, A. speB Genosensor for Rapid Detection of Streptococcus pyogenes Causing Damage of Heart Valves in Human. Cell. Mol. Biol. 2016, 62, 4. [Google Scholar] [CrossRef]
- Singh, S.; Kaushal, A.; Gautam, H.; Gupta, S.; Kumar, A. Ultrasensitive nanohybrid DNA sensor for detection of pathogen to prevent damage of heart valves. Sens. Actuators B Chem. 2017, 246, 300–304. [Google Scholar] [CrossRef]
- Ahmed, A.; Rushworth, J.V.; Wright, J.D.; Millner, P.A. Novel Impedimetric Immunosensor for Detection of Pathogenic Bacteria Streptococcus pyogenes in Human Saliva. Anal. Chem. 2013, 85, 12118–12125. [Google Scholar] [CrossRef] [PubMed]
- Coleman, H.N.; Daines, D.A.; Jarisch, J.; Smith, A.L. Chemically Defined Media for Growth of Haemophilus influenzae Strains. J. Clin. Microbiol. 2003, 41, 4408–4410. [Google Scholar] [CrossRef] [Green Version]




| SAMPLE | EOX/mV | ERED/mV | ΔE/mV | Re/Ω | CPE/µFΩ−1sn | n | Rct/Ω |
|---|---|---|---|---|---|---|---|
| bare Au | 202 | 108 | 94 | 148 | 41.4 | 0.492 | 190 |
| Au/4-ATP | 201 | 98 | 103 | 153 | 4.1 | 0.704 | 256 |
| Au/4-ATP/anti-Spy | 237 | 72 | 165 | 161 | 2.9 | 0.715 | 446 |
| Au/4-ATP/anti-Spy/BSA | 285 | 52 | 233 | 191 | 1.01 | 0.862 | 1790 |
| Method | Target Analyte | Linearity Range | LOD | Year | Ref |
|---|---|---|---|---|---|
| piezoelectric | bacterial cell | 3 × 102–3 × 106 cfu/mL | 12 cfu/mL | 2014 | 27 |
| DPV | ssG-DNA | 10−3–10−1 ng/6 μL | 130 fg/6 μL | 2017 | 29 |
| DPV | ssG-DNA | 0–1 ng/6 μL | 0.01 ng | 2014 | 30 |
| CV | ssG-DNA | 0.5–50 ng/6 μL | 0.01 ng/6 µL | 2017 | 33 |
| CV | ssG-DNA | 0–7.5 ng/6 µL | 0.10 ng/6 µL | 2016 | 35 |
| EIS | bacterial cell | 100–105 cells/10 μL | 100 cells/10 μl | 2013 | 37 |
| EIS | bacterial cell | 4.2 × 102–4.2 × 106 cfu/mL | 9.3 cfu/mL | 2020 | This work |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malinowska, N.; Białobrzeska, W.; Łęga, T.; Pałka, K.; Dziąbowska, K.; Żołędowska, S.; Czaczyk, E.; Pala, K.; Nidzworski, D. Antibody Modified Gold Electrode as an Impedimetric Biosensor for the Detection of Streptococcus pyogenes. Sensors 2020, 20, 5324. https://doi.org/10.3390/s20185324
Malinowska N, Białobrzeska W, Łęga T, Pałka K, Dziąbowska K, Żołędowska S, Czaczyk E, Pala K, Nidzworski D. Antibody Modified Gold Electrode as an Impedimetric Biosensor for the Detection of Streptococcus pyogenes. Sensors. 2020; 20(18):5324. https://doi.org/10.3390/s20185324
Chicago/Turabian StyleMalinowska, Natalia, Wioleta Białobrzeska, Tomasz Łęga, Katarzyna Pałka, Karolina Dziąbowska, Sabina Żołędowska, Elżbieta Czaczyk, Katarzyna Pala, and Dawid Nidzworski. 2020. "Antibody Modified Gold Electrode as an Impedimetric Biosensor for the Detection of Streptococcus pyogenes" Sensors 20, no. 18: 5324. https://doi.org/10.3390/s20185324