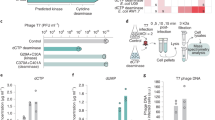
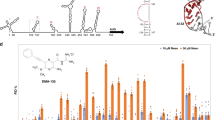
Abstract
Viperin is an interferon-induced cellular protein that is conserved in animals1. It has previously been shown to inhibit the replication of multiple viruses by producing the ribonucleotide 3′-deoxy-3′,4′-didehydro (ddh)-cytidine triphosphate (ddhCTP), which acts as a chain terminator for viral RNA polymerase2. Here we show that eukaryotic viperin originated from a clade of bacterial and archaeal proteins that protect against phage infection. Prokaryotic viperins produce a set of modified ribonucleotides that include ddhCTP, ddh-guanosine triphosphate (ddhGTP) and ddh-uridine triphosphate (ddhUTP). We further show that prokaryotic viperins protect against T7 phage infection by inhibiting viral polymerase-dependent transcription, suggesting that it has an antiviral mechanism of action similar to that of animal viperin. Our results reveal a class of potential natural antiviral compounds produced by bacterial immune systems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data that support the findings of this study are available within the article and its Extended Data and Supplementary Tables. IMG accessions, protein sequences and nucleotide sequences when relevant to the pVips appear in the Methods and Supplementary Table 2. Mass spectrometry data are available on Metabolights under study number MTBLS1750.
References
Rivera-Serrano, E. E. et al. Viperin reveals its true function. Annu. Rev. Virol. 7, 1–26 (2020).
Gizzi, A. S. et al. A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 558, 610–614 (2018).
Helbig, K. J. & Beard, M. R. The role of viperin in the innate antiviral response. J. Mol. Biol. 426, 1210–1219 (2014).
Fenwick, M. K., Li, Y., Cresswell, P., Modis, Y. & Ealick, S. E. Structural studies of viperin, an antiviral radical SAM enzyme. Proc. Natl Acad. Sci. USA 114, 6806–6811 (2017).
Fenwick, M. K., Su, D., Dong, M., Lin, H. & Ealick, S. E. Structural basis of the substrate selectivity of viperin. Biochemistry 59, 652–662 (2020).
Honarmand Ebrahimi, K., Rowbotham, J. S., McCullagh, J. & James, W. S. Mechanism of diol dehydration by a promiscuous radical-SAM enzyme homologue of the antiviral enzyme viperin (RSAD2). ChemBioChem 21, 1605–1612 (2020).
Makarova, K. S., Wolf, Y. I., Snir, S. & Koonin, E. V. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol. 193, 6039–6056 (2011).
Doron, S. et al. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359, eaar4120 (2018).
Cohen, D. et al. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 574, 691–695 (2019).
Kambara, H. et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. 42, 10668–10680 (2014).
Thiem, J. & Stangier, P. Preparative-enzymatic formation of cytidine 5′-monophosphosialate by integrated cytidine 5′-triphosphate regeneration. Liebigs Ann. Chem.1101–1105 (1990).
Dukhovny, A., Shlomai, A. & Sklan, E. H. The antiviral protein Viperin suppresses T7 promoter dependent RNA synthesis—possible implications for its antiviral activity. Sci. Rep. 8, 8100 (2018).
Makarova, K. S. et al. An updated evolutionary classification of CRISPR–cas systems. Nat. Rev. Microbiol. 13, 722–736 (2015).
Tock, M. R. & Dryden, D. T. F. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 8, 466–472 (2005).
Calendar, R. & Abedon, S. T. The Bacteriophages (Oxford Univ. Press, 2005).
Ebrahimi, K. H. et al. Viperin, through its radical-SAM activity, depletes cellular nucleotide pools and interferes with mitochondrial metabolism to inhibit viral replication. FEBS Lett. 594, 1624–1630 (2020).
Kronheim, S. et al. A chemical defence against phage infection. Nature 564, 283–286 (2018).
De Clercq, E. & Neyts, J. Antiviral Strategies (eds H. –G. Kräusslich & R. Bartenschlager) 53–84 (2009).
Feld, J. J. et al. Sofosbuvir and velpatasvir for HCV genotype 1, 2, 4, 5, and 6 infection. N. Engl. J. Med. 373, 2599–2607 (2015).
Chen, I. A. et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Steinegger, M. & Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44 (W1), W242–W245 (2016).
Eddy, S. R. Accelerated profile HMM searches. PLOS Comput. Biol. 7, e1002195 (2011).
Zimmermann, L. et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006).
Roche, B. et al. Reprint of: Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim. Biophys. Acta 1827, 923–937 (2013).
Schwartz, C. J. et al. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe–S cluster assembly proteins. Proc. Natl Acad. Sci. USA 98, 14895–14900 (2001).
Jiang, D. et al. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 82, 1665–1678 (2008).
Fortier, L.-C. & Moineau, S. in Bacteriophages: Methods and Protocols Volume 1: Isolation, Characterization, and Interactions (eds Clokie, M. R. J. & Kropinski, A.) 203–219 (Springer, 2009).
Kropinski, A. M., Mazzocco, A., Waddell, T. E., Lingohr, E. & Johnson, R. P. in Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions (eds Clokie, M. R. J. & Kropinski, A.) 69–76 (Springer, 2009).
Hsiao, J. J., Potter, O. G., Chu, T. W. & Yin, H. Improved LC/MS methods for the analysis of metal-sensitive analytes using medronic acid as a mobile phase additive. Anal. Chem. 90, 9457–9464 (2018).
Dar, D. et al. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 352, aad9822 (2016).
Acknowledgements
We thank M. Danielsen and D. Malheiro from MS-omics for conducting the MS experiments and for the extensive help with the data analysis and the Sorek laboratory members for comments on earlier versions of this manuscript. A.B. is the recipient of a European Molecular Biology Organization Long-Term Fellowship (EMBO ALTF 186-2018). A.M. was supported by a fellowship from the Ariane de Rothschild Women Doctoral Program and, in part, by the Israeli Council for Higher Education via the Weizmann Data Science Research Center. G.O. was supported by the Weizmann Sustainability and Energy Research Initiative (SAERI) doctoral fellowship. R.S. was supported, in part, by the Israel Science Foundation (personal grant 1360/16), the European Research Council (grant ERC-CoG 681203), the Ernest and Bonnie Beutler Research Program of Excellence in Genomic Medicine, the Minerva Foundation with funding from the Federal German Ministry for Education and Research, the Ben B. and Joyce E. Eisenberg Foundation and the Knell Family Center for Microbiology.
Author information
Authors and Affiliations
Contributions
A.B. and R.S. led the study and A.B. performed all experiments unless otherwise indicated. A.M. and A.B performed the computational analyses that appear in Figs. 1, 2, Extended Data Fig 9. H.S., M.M.R. and N.T. designed and performed purification of pVips and in vitro enzymatic assays that appear in Fig. 3, Extended Data Fig 6. G.M., C.A. and S.M. assisted with the plaque assays that appear in Extended Data Fig. 1. C.A. assisted in the preparation of cell lysates that appear in Fig. 3, Extended Data Figs 3–5. G.O. and G.A. assisted in the design and analysis of GFP-reporter studies presented in Fig 4, Extended Data Fig 7. R.S. supervised the study. R.S and A.B. wrote the paper together with the team.
Corresponding author
Ethics declarations
Competing interests
R.S. is a scientific cofounder and advisor of BiomX, Pantheon Bioscience and Ecophage. A.B., A.M. and R.S. are inventors on patent application PCT/IL2020/050377 licensed to Pantheon Bioscience. H.S., M.R. and N.T. are employed by Pantheon Bioscience. The other authors have no competing interests.
Additional information
Peer review information Nature thanks Pieter Dorrestein, Pei-Yong Shi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Fig. 1 pVips protect against phage infection.
Bacteria expressing pVips, GFP or MoaA (negative controls), or the human viperin gene were grown on agar plates and tenfold serial dilutions of the phage lysate were dropped on the plates. a–h, Efficiency of plating (EOP) data, representing plaque-forming units per millilitre; each bar graph represents average of three replicates, with individual data points overlaid.
Extended Data Fig. 2 T7 infection in liquid culture in the presence of pVips.
a, For each pVip, growth curves of liquid cultures infected by phage T7 (MOI 0.001) are shown. Light and dark grey are uninfected and infected controls (strain expressing GFP), respectively. Light and dark red are uninfected and infected strains expressing pVips, respectively. Two technical replicates are presented as individual curves; representative of three biological replicates. The negative controls (GFP uninfected, GFP infected) are the same for pVips 6, 7, 8, 10, 15, 27, 37, 39, 42, 50, 54, MoaA, and for pVip12, 19, 32, 44, 46, 47, 48, 57, 58, 60, 61, 62, 63. b, The catalytic activity of pVips is required for defence against T7 phage. For each pVip and its respective mutant (mutation of three cysteines in the active site), growth curves of liquid cultures infected by phage T7 (MOI 0.001) are presented. Light and dark grey are uninfected and infected controls (strain expressing MoaA), respectively. Light and dark red are uninfected and infected strains expressing viperins, respectively. Light and dark blue are uninfected and infected strains expressing catalytically inactive mutants. Two technical replicates are presented as individual curves; representative of three biological replicates.
Extended Data Fig. 3 Detection of ddhCTP and ddhCTP derivatives in cell lysates from an E. coli strain expressing the human viperin.
a, Extracted ion chromatogram of the ddhC standard. b–d, Extracted ion chromatogram for singly charged masses that are predicted to correspond to ddhC (m/z 226.0822, retention time (RT) of 2.2 min) (b), ddhCMP (m/z 306.0486, RT 9.7) (c) and ddhCTP (m/z 465.9812, RT 10.7) (d) in cell lysates from an E. coli strain expressing the human viperin. Representative of three replicates.
Extended Data Fig. 4 Detection of ddh-ribonucleotides in lysates of cells that express pVips.
a, Quantification of ddh-cytidine (ddhC) in lysates of cells expressing pVips. Detection and quantification of ddhC was performed using LC–MS with a synthesized chemical standard (Methods). For MoaA, the measurement was under the limit of detection (LOD 0.0003 uM). Bar graph represents average of three replicates, with individual data points overlaid. b–h, Relative abundance for singly charged masses that are predicted to correspond to ddhC (m/z 226.0822, retention time (RT) of 2.2 min) (b), ddhCMP (m/z 306.0486, RT 9.7) (c), ddhCTP (m/z 465.9812, RT 10.7) (d), ddhUMP (m/z 307.0326, RT 8.7) (e), ddhUTP (m/z 466.9652, RT 9.9) (f), ddhGMP (m/z 346.0547, RT 9.8) (g), and ddhGTP (m/z 505.9874, RT 10.7) (h). Average relative abundance is presented as bar graph, with individual data points from three biological replicates overlaid. Limit of detection (LOD) is indicated by a dashed grey line. A compound was defined as present, in Fig. 3, if all three replicates were above the LOD.
Extended Data Fig. 5 MS/MS fragmentation spectra for predicted compounds.
a–c, MS/MS data were acquired in positive ionization mode for a synthesized chemical standard ddhC (a) as well as for masses from the human viperin cell lysate predicted to correspond to ddhC (b), and ddhCMP (c). d, e, Similar data were obtained for masses from the pVip21 cell lysate predicted to correspond to ddhGMP (d), and ddhGTP (e). f, g, MS/MS data were acquired, in negative ionization mode, from the pVip47 cell lysate for masses predicted to correspond to ddhUMP (f), and ddhUTP (g). In all panels, assignment of hypothetical structures is indicated for informative fragment ions. The ddhC molecule is annotated to level 1, and all other molecules are annotated to level 2b, per the Metabolomics Standards Initiative nomenclature.
Extended Data Fig. 6 MS/MS fragmentation spectra for predicted compounds from in vitro reactions with purified pVips.
a, b, MS/MS data were acquired in positive ionization mode for the product detected in reaction samples using purified pVip6 or purified pVip56 and CTP and GTP as nucleotide substrates respectively; the resulting products are predicted to correspond to ddhCTP (a) and ddhGTP (b). c, MS/MS data were acquired in negative ionization mode for product detected in reaction samples using purified pVip8 UTP as substrate; the resulting product is predicted to correspond to ddhUTP.
Extended Data Fig. 7 Transcription during induction of WT and mutant pVips.
a, The catalytic activity of pVips is required for defence against T7 phage and repression of viral transcription. Application of the reporter assay (same as presented in Fig. 4a) for strains expressing the human viperin, pVips and their cognate catalytically inactive mutants. Strains are first induced with arabinose for 45 min to express the pVip. At t = 0, IPTG is added to express the GFP. Fluorescence/OD over time curves are presented for each strain. Dark and light red correspond to induced and non-induced wild type viperins, respectively; Dark and light blue correspond to induced and non-induced mutant viperins, respectively. Grey curve corresponds to negative control (WT viperin, no addition of IPTG). Two technical replicates are presented as individual curves. Representative of two biological replicates. b, T7 RNAP expression as measured by RNA-seq. The expression (RPKM) of T7 RNAP in cells expressing viperins was compared to that in cells expressing the MoaA negative control. Bar graphs represent average of two replicates, with individual data points overlaid.
Extended Data Fig. 8 Heterologous expression of pVips is not toxic is E. coli.
Expression of pVips, human viperin or negative controls (GFP, MoaA) was induced at 45 min by addition of arabinose (final concentration 0.2%). Colony forming units (CFU) were measured right after dilution from overnight culture (t = 0), before induction (t = 45), and 45 and 90 min after induction (t = 90, t = 135). Bar graphs represent average of three replicates, with individual data points overlaid.
Extended Data Fig. 9 Putative multi-gene defence systems that include pVips.
Representative instances of pVips and their genomic neighbourhood. Genes predicted to be part of the pVip-containing defence system are highlighted. Genes known to be involved in defence are in yellow. Genes of mobile genetic elements are in dark grey. RM, restriction-modification; TA, toxin-antitoxin. The name of bacterial species, and the accession of the relevant genomic scaffold in the IMG database20 are indicated on the left. a–d, Four common configurations of putative pVip-containing systems found in bacterial and archaeal genomes.
Extended Data Fig. 10 Phylogenetic tree of pVips and putative eukaryotic viperins.
MoaA sequences were used as an outgroup (grey). pVips are depicted in red and putative eukaryotic viperins selected for the phylogenetic tree presented in Fig. 2 are depicted in blue.
Supplementary information
Supplementary Table 1
Clusters of genes retrieved by the homology-based search of human viperin in prokaryotic genomes. Genes used to calculate defense scores were those present on DNA scaffolds of sufficient size with at least ten genes from each side of the viperin homolog.
Supplementary Table 2
A list of pVips. Gene and genome accessions in the IMG database are indicated.
Supplementary Table 3
MoaA and eukaryotic viperin sequences. MoaA genes (outgroup) and eukaryotic viperin sequences used in the phylogenetic tree in Figure 2.
Supplementary Table 4
A list of primers used in this study.
Supplementary Table 5
A list of phages used in this study.
Rights and permissions
About this article
Cite this article
Bernheim, A., Millman, A., Ofir, G. et al. Prokaryotic viperins produce diverse antiviral molecules. Nature 589, 120–124 (2021). https://doi.org/10.1038/s41586-020-2762-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2762-2
This article is cited by
-
Conservation and similarity of bacterial and eukaryotic innate immunity
Nature Reviews Microbiology (2024)
-
Phages overcome bacterial immunity via diverse anti-defence proteins
Nature (2024)
-
Structures and activation mechanism of the Gabija anti-phage system
Nature (2024)
-
Bacterial defences: mechanisms, evolution and antimicrobial resistance
Nature Reviews Microbiology (2023)
-
Origins and diversification of animal innate immune responses against viral infections
Nature Ecology & Evolution (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.