Abstract
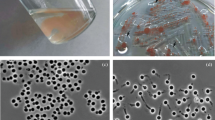
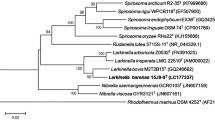
A novel strain of Planctomycetes, designated JC670T, was isolated from a high altitude (~ 2900 m above sea level) soil sample collected from Garhwal region in the Western Himalaya. Colonies of this strain were observed to be light pink coloured with spherical to oval shaped cells having crateriform structures distributed all over the cell surface. The cells divide by budding. Strain JC670T was found to grow well at pH 7.0 and pH 8.0 and to tolerate up to 2% NaCl (w/v). MK6 was the only respiratory quinone identified. The major fatty acids of strain JC670T were identified as C18:1ω9c, C18:0 and C16:0, and phosphatidylcholine, two unidentified phospholipids and six unidentified lipids are present as the polar lipids. The polyamines putrescine and sym-homospermidine were detected. Strain JC670T shows high 16S rRNA gene sequence identity (95.4%) with Paludisphaera borealis PX4T. The draft genome size of strain JC670T is 7.97 Mb, with G + C content of 70.4 mol%. Based on phylogenetic analyses with the sequences of ninety-two core genes, low dDDH value (20.6%), low gANI (76.8%) and low AAI (69.1%) results, differential chemotaxonomic and physiological properties, strain JC670T (= KCTC 72850T = NBRC 114339T) is recognised as the type strain of a new species of the genus Paludisphaera, for which we propose the name Paludisphaera soli sp. nov.



Similar content being viewed by others
Abbreviations
- NCBI:
-
National Center for Biotechnology Information
- ANI:
-
Average Nucleotide Index
- AAI:
-
Average Nucleotide Index
- dDDH:
-
Digital DNA-DNA Hybridization
- HPLC:
-
High-Pressure Liquid Chromatography
- KCTC:
-
Korean Collection for Type Cultures
References
Auch AF, Klenk HP, Göker M (2010) Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci 2:142–148
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M et al (2008) The RAST server: rapid annotations using subsystems technology. BMC genomics 9:75
Baghel VS, Tripathi RD, Ramteke PW, Gopal K, Dwivedi S, Jain RK, Rai UN, Singh SN (2005) Psychrotrophic proteolytic bacteria from cold environment of Gangotri glacier, Western Himalaya, India. Enzyme Microb Tech 36:654–659
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87
Bondoso J, Albuquerque L, Nobre MF, Lobo-da-Cunha A, da Costa MS, Lage OM (2011) Aquisphaera giovannonii gen. nov., sp. nov., a planctomycete isolated from a freshwater aquarium. Int J Syst Evol Microbiol 61:2844–2850
Bondoso J, Albuquerque L, Nobre MF, Lobo-da-Cunha A, da Costa MS, Lage OM (2015) Roseimaritima ulvae gen. nov., sp. nov. and Rubripirellula obstinate gen. nov., sp. nov. two novel planctomycetes isolated from the epiphytic community of macroalgae. Syst Appl Microbiol 38:8–15
Borneman J, Skroch PW, O'Sullivan KM, Palus JA, Rumjanek NG, Jansen JL, Nienhuis J, Triplett EW (1996) Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol 62:1935–1943
Borneman J, Triplett EW (1997) Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol 63:2647–2653
Buckley DH, Huangyutitham V, Nelson TA, Rumberger A, Thies JE (2006) Diversity of planctomycetes in soil in relation to soil history and environmental heterogeneity. Appl Environ Microbiol 72:4522–4531
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, Meyer SD, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Darzi Y, Letunic I, Bork P, Yamada T (2018) iPath3. 0: interactive pathways explorer v3. Nucleic Acids Res 46:W510–W513
Dedysh SN, Ivanova AA (2019) Planctomycetes in boreal and subarctic wetlands: diversity patterns and potential ecological functions. FEMS Microbiol Eco 95:fiy227
Dedysh SN, Kulichevskaya IS, Beletsky AV, Ivanova AA, Rijpstra WIC, Damsté JSS, Ravin NV (2020) Lacipirellula parvulagen. nov., sp. nov., representing a lineage of planctomycetes widespread in low-oxygen habitats, description of the family Lacipirellulaceae fam. nov. and proposal of the orders Pirellulalesord. nov., Gemmatalesord. nov. and Isosphaerales ord. nov. Syst Appl Microbiol 43:126050
Eisenreich W, Bacher A, Arigoni D, Rohdich F (2004) Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell Mol Life Sci 61:1401–1426
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Imhoff JF (1984) Quinones of phototrophic purple bacteria. FEMS Microbiol Lett 25:85–89
Ivanova AO, Dedysh SN (2012) Abundance, diversity, and depth distribution of planctomycetes in acidic northern wetlands. Front Microbiol 3:5
Ivanova AA, Naumoff DG, Miroshnikov KK, Liesack W, Dedysh SN (2017) Comparative genomics of four Isosphaeraceae planctomycetes: a common pool of plasmids and glycoside hydrolase genes shared by Paludisphaera borealis PX4T, Isosphaera pallida IS1BT, Singulisphaera acidiphila DSM 18658T, and strain SH-PL62. Front Microbiol 8:412
Jogler C, Wiegand S, Boedeker C, Heuer A, Peeters SH, Jogler M, Jetten MSM, Rohde M, Kallscheuer N (2020) Tautonia plasticadhaerens sp. nov., a novel species in the family Isosphaeraceae isolated from an alga in a hydrothermal area of the Eolian Archipelago. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-020-01424-3
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9
Kates M (1972) Isolation, analysis and identification of lipids. Tech Lipidol 3:268–618
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Köhler T, Stingl U, Meuser K, Brune A (2008) Novel lineages of Planctomycetes densely colonize the alkaline gut of soil-feeding termites (Cubitermes spp.). Environ Microbiol 10:1260–1270
Kulichevskaya IS, Ivanova AA, Suzina NE, Rijpstra WIC, Damste JSS, Dedysh SN (2016) Paludisphaera borealis gen. nov., sp. nov., a hydrolytic planctomycete from northern wetlands, and proposal of Isosphaeraceae fam. nov. Int J Syst Evol Microbiol 66:837–844
Kumar D, Gaurav K, Uppada J, Deepshikha G, Ch S, Ch VR (2020a) Roseimaritima sediminicola sp. nov., a new member of Planctomycetaceae isolated from Chilika lagoon. Int J Syst Evol Microbiol 70:2616–2623
Kumar D, Gaurav K, Sreya PK, Shabbir A, Uppada J, Sasikala Ch, Ramana CV (2020b) Gimesia chilikensis sp. nov., a haloalkali-tolerant planctomycete isolated from Chilika lagoon and emended description of the genus Gimesia. Int J Syst Evol Microbiol 70:3647–3655
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kumar S, Suyal DC, Yadav A, Shouche Y, Goel R (2019) Microbial diversity and soil physiochemical characteristic of higher altitude. PLoS ONE 14:e0213844
Mac Faddin JF (1985) Media for isolation-cultivation-identification-maintenance of medical bacteria. Williams & Wilkins, Philadelphia
Mc Inerney JO, Martin WF, Koonin EV, Allen JF, Galperin MY, Lane N, Embley AJM, TM, (2011) Planctomycetes and eukaryotes: a case of analogy not homology. BioEssays 33:810–817
Meier-Kolthoff JP, Klenk HP, Göker M (2014) Taxonomic use of DNA G+ C content and DNA–DNA hybridization in the genomic age. Int J Syst Evol Microbiol 64:352–356
Na SI, Kim YO, Yoon SH, Ha SM, Baek I, Chun J (2018) UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:280–285
Neef A, Amann R, Schlesner H, Schleifer KH (1998) Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257–3266
Oren A, Duker S, Ritter S (1996) The polar lipid composition of Walsby's square bacterium. FEMS Microbiol Lett 138:135–140
Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou J, Oren A, Zhang YZ (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol Res 196:2210–2215
Rodriguez RLM, Konstantinidis KT (2014) Bypassing cultivation to identify bacterial species. Microbe 9:111–118
Rodriguez-R LM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes (No. e1900v1). Peer J Preprints
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids.
Stackebrandt E, Liesack W, Goebel BM (1993) Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J 7:232–236
Wagner M, Horn M (2006) The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17:241–249
Wang J, Jenkins C, Webb RI, Fuerst JA (2002) Isolation of Gemmata-like and Isosphaera-like planctomycete bacteria from soil and freshwater. Appl Environ Microbiol 68:417–422
Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW et al (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45:D535–D542
Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput Biol 13:e1005595
Wiegand S, Jogler M, Jogler C (2018) On the maverick Planctomycetes. FEMS Microbiol Rev 42:739–760
Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, Oberbeckmann S et al (2020) Cultivation and functional characterization of 79 Planctomycetes uncovers their unique biology. Nat Microbiol 5:126–140
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613
Zhao L, Chang WC, Xiao Y, Liu HW, Liu P (2013) Methylerythritol phosphate pathway of isoprenoid biosynthesis. Annu Rev Biochem 82:497–530
Acknowledgements
MS thanks CSIR, RK and KG thank the DBT, New Delhi and JU thanks TEQIP III for the award for research fellowships. Ramana thanks the Department of Biotechnology, Government of India for the award of Tata Innovation fellowship. Sasikala thanks UGC for Mid-Career Award. We thank the staff and the laboratory facilities provided by Centre for Interdisciplinary Studies of Mountain & Hill Environment, University of Delhi.
Funding
This work received grant specifically from Department of Biotechnology, Government of India. Financial support received from Council for Scientific and Industrial Research (CSIR), and AICTE, New Delhi is acknowledged. Infrastructure facilities funded by DST-FIST, UGC-SAP (DRS) and TEQIP, AICTE-RPS and MODROBS are acknowledged. The financial assistance of DST-PURSE Grant to MKP is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
RK and MS performed sample collection. RK, MS and KG isolated the strain and performed the initial cultivation, strain deposition and strain characterisation, KG performed the electron microscopic analysis and media optimisation, JU performed the genomic and phylogenetic analysis, SA performed and analysed the data for polyamines, RK, MS and KG wrote the manuscript, Ramana, Sasikala and MKP supervised the study and contributed to text preparation and revised the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kaushik, R., Sharma, M., Gaurav, K. et al. Paludisphaera soli sp. nov., a new member of the family Isosphaeraceae isolated from high altitude soil in the Western Himalaya. Antonie van Leeuwenhoek 113, 1663–1674 (2020). https://doi.org/10.1007/s10482-020-01471-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-020-01471-w