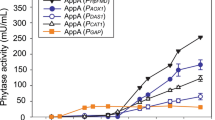
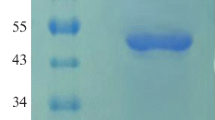
Abstract
Phytase is an additive in animal feed that degrades phytic acid in plant material, reducing feeding costs, and pollution from fecal phosphorus excretion. A multistrategy approach was adopted to improve the expression of E. coli phytase in Pichia pastoris. We determined that the most suitable signal peptide for phytase secretion was an α-factor secretion signal with an initial enzyme activity of 153.51 U/mL. Increasing the copy number of this gene to four increased phytase enzyme activity by 234.35%. PDI overexpression and Pep4 gene knockout increased extracellular phytase production by 35.33% and 26.64%, respectively. By combining favorable factors affecting phytase expression and secretion, the enzyme activity of the phytase-engineered strain was amplified 384.60% compared with that of the original strain. We also evaluated the potential for the industrial production of the engineered strain using a 50-L fed-batch fermenter and achieved a total activity of 30,246 U/mL after 180 h of fermentation.





Similar content being viewed by others
References
Vasudevan UM, Krishna S, Jalaja V et al (2017) Microbial phytase: impact of advances in genetic engineering in revolutionizing its properties and applications. Biores Technol 245:1790–1799. https://doi.org/10.1016/j.biortech.2017.05.060
Reddy NR, Sathe SK, Salunkhe DK (1982) Phytates in legumes and cereals. Adv Food Res 28:1–92. https://doi.org/10.1016/s0065-2628(08)60110-x
Lei XG, Weaver JD, Mullaney E et al (2013) Phytase, a new life for an “old” enzyme. Annu Rev Anim Biosci 1:283–309. https://doi.org/10.1146/annurev-animal-031412-103717
Kebreab E, Hansen AV, Strathe AB (2012) Animal production for efficient phosphate utilization: from optimized feed to high efficiency livestock. Curr Opin Biotech 23:872–877. https://doi.org/10.1016/j.copbio.2012.06.001
Jain J, Kumar A, Singh D et al (2018) Purification and kinetics of a protease-resistant, neutral, and thermostable phytase from Bacillus subtilis subspsubtilis JJBS250 ameliorating food nutrition. Prep Biochem Biotech 48:8. https://doi.org/10.1080/10826068.2018.1487848
Jain J, Singh B (2017) Phytase production and development of an ideal dephytinization process for amelioration of food nutrition using microbial phytases. Appl Biochem Biotech 181:1485–1495. https://doi.org/10.1007/s12010-016-2297-z
Kim OH, Kim YO, Shim JH et al (2010) β-propeller phytase hydrolyzes insoluble Ca2+-phytate salts and completely abrogates the ability of phytate to chelate metal ions. Biochemistry 49:10216–10227. https://doi.org/10.1021/bi1010249
Leytem AB, Widyaratne GP, Thacker PA (2014) Phosphorus utilization and characterization of ileal digesta and excreta from broiler chickens fed diets varying in cereal grain, phosphorus level, and phytase addition. Poultry Sci 87:2466–2476. https://doi.org/10.3382/ps.2008-00043
Pakbaten B, Heravi RM, Kermanshahi H et al (2019) Production of phytase enzyme by a bioengineered probiotic for degrading of phytate phosphorus in the digestive tract of poultry. Probiotics Antimicrob 11:580–587. https://doi.org/10.1007/s12602-018-9423-x
Tai HM, Yin LJ, Chen WC et al (2013) Overexpression of Escherichia coli phytase in Pichia pastoris and its biochemical properties. J Agric Food Chem 61:6007–6015. https://doi.org/10.1021/jf401853b
Lu H, Kühn I, Bedford MR et al (2019) Effect of phytase on intestinal phytate breakdown, plasma inositol concentrations and glucose transporter type 4 abundance in muscle membranes of weanling pigs. J Anim Sci 97:3907–3919. https://doi.org/10.1093/jas/skz234
Augspurger N, Webel D, Lei X et al (2003) Efficacy of an E. coli phytase expressed in yeast for releasing phytate-bound phosphorus in young chicks and pigs. J Anim Sci 81:474–483. https://doi.org/10.2527/2003.812474x
Luo HY, Yao B et al (2004) Overexpression of Escherchia coli phytase with high specific activity. Sheng Wu Gong Cheng Xue Bao 20(1):78–84. https://doi.org/10.13345/j.cjb.2004.01.018
Luo HY, Huang HQ, Bai YG et al (2006) Improving phytase expression by increasing the gene copy number of appA-m in Pichia pastoris. Sheng Wu Gong Cheng Xue Bao 22(4):528–533. https://doi.org/10.1016/s1872-2075(06)60041-1
Rodriguez E, Wood ZA et al (2000) Site-directed mutagenesis improves catalytic efficiency and thermostability of Escherichia coli pH 2.5 acid phosphatase/phytase expressed in Pichia pastoris. Arch Biochem Biophys 382(1):105–112. https://doi.org/10.1006/abbi.2000.2021
Zhang WH, Liu CP, Inan M et al (2004) Optimization of cell density and dilution rate in Pichia pastoris continuous fermentations for production of recombinant proteins. J Ind Microbiol Biot 31(7):330–334. https://doi.org/10.1007/s10295-004-0155-4
Huang MT, Bao JC, Hallström BM et al (2017) Efficient protein production by yeast requires global tuning of metabolism. Nat Commun 8(1):1131. https://doi.org/10.1038/s41467-017-00999-2
Liu ZH, Liu LF et al (2014) Improved production of a heterologous amylase in Saccharomyces cerevisiae by inverse metabolic engineering. Appl Environ Microb 80(17):5542–5550. https://doi.org/10.1128/AEM.00712-14
Hartner FS, Ruth C, Langenegger D et al (2008) Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res 36(12):e76. https://doi.org/10.1093/nar/gkn369
Cregg JM, Barringer KJ, Hessler AY et al (1985) Pichia pastoris as a host system for transformations. Mol Cell Biol 5(12):3376–3385
Litzinger S, Duckworth A, Nitzsche K et al (2010) Muropeptide rescue in Bacillus subtilis involves sequential hydrolysis by beta-N-Acetylglucosaminidase and N-Acetylmuramyl-l-Alanine amidase. J Bacteriol 192(12):3132–3143. https://doi.org/10.1128/JB.01256-09
Jiang S, Jiang HY, Zhou YL et al (2019) High-level expression of β-N-Acetylglucosaminidase BsNagZ in Pichia pastoris to obtain GlcNAc. Bioproc Biosyst Eng 42(4):611–619. https://doi.org/10.1007/s00449-018-02067-5
Whyteside G, Alcocer MJC, Kumita JR et al (2011) Native-state stability determines the extent of degradation relative to secretion of protein variants from Pichia pastoris. PLoS ONE 6(7):e22692. https://doi.org/10.1371/journal.pone.0022692
Sakabe I, Hu R, Jin L et al (2015) TMEM33: a new stress-inducible endoplasmic reticulum transmembrane protein and modulator of the unfolded protein response signaling. Breast Cancer Res Treat 153:285–297. https://doi.org/10.1007/s10549-015-3536-7
Damasceno LM, Huang CJ et al (2012) Protein secretion in Pichia pastoris and advances in protein production. Appl Microbiol Biotechnol 93:31–39. https://doi.org/10.1007/s00253-011-3654-z
Guerfal M, Ryckaert S, Jacobs PP et al (2010) Research the HAC1 gene from Pichia pastoris: characterization and effect of its overexpression on the production of secreted surface displayed and membrane proteins. Microb Cell Fact 9:49. https://doi.org/10.1186/1475-2859-9-49
Lin XQ, Han SY, Zhang N et al (2013) Bleach boosting effect of xylanase A from Bacillus halodurans C-125 in ECF bleaching of wheat straw pulp. Enzyme Microb Tech 52:91–98. https://doi.org/10.1016/j.enzmictec.2012.10.011
Kim MD, Han KC et al (2003) Coexpression of BiP increased antithrombotic hirudin production in recombinant Saccharomyces cerevisiae. J Biotechnol 101:81–87. https://doi.org/10.1016/s0168-1656(02)00288-2
Shusta EV, Raines RT, Plückthun A et al (1998) Increasing the secretory capacity of Saccharomyces cerevisiae for production of single-chain antibody fragments. Nat Biotechnol 16(8):773–777. https://doi.org/10.1038/nbt0898-773
Seo M, Ryou HJ, Yun EY et al (2015) Molecular characterization of endoplasmic reticulum oxidoreductin 1 from bombyx mori. Int J Mol Sci 16:26520–26529. https://doi.org/10.3390/ijms161125977
Li JD, Li XL, Gai YM et al (2019) Evolution of E. coli phytase for increased thermostability guided by rational parameters. J Microbiol Biotechnol 29(3):419–428. https://doi.org/10.4014/jmb.1811.11017
Zhang W, Wang H, Zhang L et al (2011) Large-scale assignment of N-glycosylation sites using complementary enzymatic deglycosylation. Talanta 85(1):499–505. https://doi.org/10.1016/j.talanta.2011.04.019
Li C, Lin Y, Zheng X et al (2015) Combined strategies for improving expression of Citrobacter amalonaticus phytase in Pichia pastoris. BMC Biotechnol 15:88. https://doi.org/10.1186/s12896-015-0204-2
Hryhorowicz M, Zeyland J et al (2017) CRISPR/Cas9 immune system as a tool for genome engineering. Arch Immunol 65:233–240. https://doi.org/10.1007/s00005-016-0427-5
Fonseca-Maldonado R, Vieira DS, Alponti JS et al (2013) Engineering the pattern of protein glycosylation modulates the thermostability of a GH11 xylanase. J Biol Chem 288(35):25522–25534. https://doi.org/10.1074/jbc.M113.485953
Shental-Bechor D, Levy Y (2008) Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc Natl Acad Sci 105(24):8256–8261. https://doi.org/10.1073/pnas.0801340105
Yao MZ, Wang X, Wang W et al (2013) Improving the thermostability of Escherichia coli phytase, appA, by enhancement of glycosylation. Biotechnol Lett 35(10):1669–1676. https://doi.org/10.1007/s10529-013-1255-x
Wu TH, Chen CC, Cheng YS et al (2014) Improving specific activity and thermostability of Escherichia coli phytase by structure-based rational design. J Biotechnol 175:1–6. https://doi.org/10.1016/j.jbiotec.2014.01.034
Ahmad M, Hirz M, Pichler H et al (2014) Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol 98:5301–5317. https://doi.org/10.1007/s00253-014-5732-5
Hohenblum H, Gasser B, Maurer M et al (2003) Effects of gene dosage, promoters, and substrates on unfolded protein stress of recombinant Pichia pastoris. Biotechnol Bioeng 85(4):367–375. https://doi.org/10.1002/bit.10904
Marsalek L, Gruber C, Altmann F et al (2017) Disruption of genes involved in CORVET complex leads to enhanced secretion of heterologous carboxylesterase only in protease deficient Pichia pastoris. Biotechnol J 12:1600584. https://doi.org/10.1002/biot.201600584
Tsai B, Rodighiero C, Lencer WI et al (2001) Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104:937–948. https://doi.org/10.1016/s0092-8674(01)00289-6
Inan M, Aryasomayajula D, Sinha J et al (2006) Enhancement of protein secretion in Pichia pastoris by overexpression of protein disulfide isomerase. Biotechnol Bioeng 93(4):771–778. https://doi.org/10.1002/bit.20762
Sha C, Yu XW, Lin NX et al (2013) Enhancement of lipase r27RCL production in Pichia pastoris by regulating gene dosage and co-expression with chaperone protein disulfide isomerase. Enzyme Microb Tech 53:438–443. https://doi.org/10.1016/j.enzmictec.2013.09.009
Daly R, Hearn MTW (2005) Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J Mol Recognit 18:119–138. https://doi.org/10.1002/jmr.687
Fernández FJ, López-Estepa M et al (2016) Production of protein complexes in Non-methylotrophic and methylotrophic yeasts. Adv Exp Med Biol 896:137–153. https://doi.org/10.1007/978-3-319-27216-0_9
Zhou XS, Zhang YX (2002) Decrease of proteolytic degradation of recombinant hirudin produced by Pichia pastoris by controlling the specific growth rate. Biotechnol Lett 24(17):1449–1453. https://doi.org/10.1023/A:1019831406141
Cho EY, Cheon SA, Kim H et al (2010) Multiple-yapsin-deficient mutant strains for high-level production of intact recombinant proteins in Saccharomyces cerevisiae. J Biotechnol 149:1–7. https://doi.org/10.1016/j.jbiotec.2010.06.014
Wu M, Shen Q, Yang Y et al (2013) Disruption of YPS1 and PEP4 genes reduces proteolytic degradation of secreted HSA/PTH in Pichia pastoris GS115. J Ind Microbiol Biotechnol 40:589–599. https://doi.org/10.1007/s10295-013-1264-8
Marques M, Mojzita D, Amorim MA (2006) The Pep4p vacuolar proteinase contributes to the turnover of oxidized proteins but PEP4 overexpression is not sufficient to increase chronological lifespan in Saccharomyces cerevisiae. Microbiology 152:3595–3605. https://doi.org/10.1099/mic.0.29040-0
Kerstens W, Dijck PV (2018) A cinderella story: how the vacuolar proteases Pep4 and Prb1 do more than cleaning up the cell’s mass degradation processes. Microb Cell 5(10):438–443. https://doi.org/10.15698/mic2018.10.650
Marsalek L, Puxbaum V et al (2019) Disruption of vacuolar protein sorting components of the HOPS complex leads to enhanced secretion of recombinant proteins in Pichia pastoris. Microb Cell Fact 18:119. https://doi.org/10.1186/s12934-019-1155-4
Horazdovsky BF, Cowles CR, Mustol P et al (1996) A novel RING finger protein, Vps8p, functionally interacts with the small GTPase, Vps21p, to facilitate soluble vacuolar protein localization. J Biol Chem 271(52):33607–33615. https://doi.org/10.1074/jbc.271.52.33607
Schutter KD, Lin YC (2009) Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol 27(6):561–566. https://doi.org/10.1038/nbt.1544
Akbarzadeh A, Dehnavi E, Aghaeepoor M et al (2015) Optimization of recombinant expression of synthetic bacterial phytase in Pichia pastoris using response surface methodology. Jundishapur J Microb 8(12):e27553. https://doi.org/10.5812/jjm.27553
Xiong AS, Yao QH, Peng RH et al (2006) High level expression of a synthetic gene encoding Peniophora lycii phytase in methylotrophic yeast Pichia pastoris. Appl Microbiol Biotechnol 72(5):1039–1047. https://doi.org/10.1007/s00253-006-0384-8
Acknowledgements
This work was supported by the National Key R&D Program of China (2018YFD0901001) and the Tianjin Science Fund for Distinguished Young Scholars (17JCJQJC45300). We thank Ms. Jie Zhang from TIB-CAS for her help with the enzyme activity determination.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Helian, Y., Gai, Y., Fang, H. et al. A multistrategy approach for improving the expression of E. coli phytase in Pichia pastoris. J Ind Microbiol Biotechnol 47, 1161–1172 (2020). https://doi.org/10.1007/s10295-020-02311-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-020-02311-6