Abstract
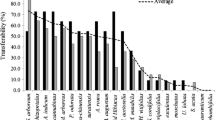
Transferability of SSR markers from related species/genera is a rapid and cost-effective method to enhance genomic database of genetically neglected crops. Monotypic genus Tecomella undulata, a pharmacologically important endangered timber tree, found in hyper arid regions is one such genetic resource limited species, wherein neither SSR identification nor cross transferability studies have been initiated. Transferability of 69 cross-genera SSR primers selected from other members of family bignoniaceae (Incarvillea sinensis, I. mairei, Jacaranda copaia, Tabebuia aurea and Arrabidaea chica) showed 40.58% (28 primers) transferability amplifying 1–10 amplicons ranging in the size from 80to 600 bp in 24 accessions of T. undulata. Per cent marker transferability varied from 66.67% (A. chica) to 38.89% (J. copaia) among the genera tested while no transferable markers could be identified from I. sinensis. Transferability of simple di-/tri-nucleotide repeat (34.2–37.5%) and complex nucleotide sequence (60.0%) based SSR motifs were higher as compared to penta-/hexa-nucleotide based repeat motifs. Within the 28 transferable markers, 26 (92.86%) were polymorphic while 2 (7.14%) were monomorphic. Average number of alleles per primer was 5.39 while average polymorphic alleles per locus were 5.11. The PIC values of SSR markers ranged from 0.23 to 0.85 with an average of 0.59. Efficiency of the identified cross species SSR markers was tested for diversity analysis using a set of 24 trees (8 each of yellow, orange and red flower colour morphotypes). The 24 samples clustered into six groups with similarity coefficient ranging from 0.29 to 0.67, while six accessions formed distinct out groups. Distribution of accessions in the SSR dendrogram showed no correlation with flower colour. More diversity (99%) was observed within populations compared to that existing between populations (1%). The present study can enrich the genomic background of T. undulata by identifying suitable polymorphic markers from confamiliar species that can be applied for genetic diversity studies, mapping of QTLs and cultivar identification as well.





Similar content being viewed by others
Abbreviations
- AFLP:
-
Amplified fragment length polymorphism
- AMOVA:
-
Analysis of molecular variance
- CTAB:
-
Cetyltrimethyl ammonium bromide
- EST:
-
Expressed sequence tags
- ISSR:
-
Inter simple sequence repeat
- ITS:
-
Internal transcribed spacer
- PCoA:
-
Principal coordinates analysis
- PIC:
-
Polymorphic information content
- RAPD:
-
Randomly amplified polymorphic DNA
- SCoT:
-
Start codon targeted polymorphism
- SSLP:
-
Simple sequence length polymorphism
- SSR:
-
Simple sequence repeats
- STR:
-
Short tandem repeats
References
Ai HL, Wang H, Li DZ, Yang JB (2009) Isolation and characterization of 13 microsatellite loci from Incarvellia mairei (Bignoniaceae), an endemic species to the Himalaya-Hengduan mountain region. Conserv Genet 10:1613–1615
Alves FM, Zucci MI, Azevedo-Tozzi AM, Sartori ALB, Souza AP (2014) Characterization of microsatellite markers developed from Prosopis rubriflora and Prosopis ruscifolia (Leguminosae-Mimosoideae), legume species that are used as models for genetic diversity studies in Chaquenian areas under anthropization in South America. BMC Res Note 7:375
Baloch FS, Alsaleh A, Shahid MQ, Çiftçi V, Sáenz LE, de Miera AM, Nadeem MA, Aktaş H, Özkan H, Hatipoğlu R (2017) A whole genome DArT seq and SNP analysis for genetic diversity assessment in durum wheat from Central fertile Crescent. PLoS ONE 12:e0167821. https://doi.org/10.1371/journal.pone.0167821
Bhau BS, Negi MS, Jindal SK, Singh M, Lakshmikumaran M (2007) Assessing genetic diversity of Tecomella undulata (Sm.)—an endangered tree species using amplified fragment length polymorphisms-based molecular markers. Curr Sci 93:67–72
Bombonato JR, Bonatelli IAS, Silva RGA, Moraes EM, Zappi DC, Taylor NP, Franco FF (2019) Cross-genera SSR transferability in cacti revealed by a case study using Cereus (Cereae, Cactaceae). Genet Mol Biol 42(1):87–94
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Braga AC, Reis AMM, Leoi LT, Pereira RW, Collevatti RG (2007) Development and characterization of microsatellite markers for the tropical tree species Tabebuia aurea (Bignoniaceae). Mol Ecol Notes 7:53–56
Chhajer S, Jukanti AK, Kalia RK (2017) Start codon targeted (SCoT) polymorphism-based genetic relationships and diversity among populations of Tecomella undulata (Sm.) Seem—an endangered timber tree of hot arid regions. Tree Genet Genom 13:84
Chhajer S, Jukanti AK, Bhatt RK, Kalia RK (2018) Genetic diversity studies in endangered desert teak [Tecomella undulata (Sm) Seem] using arbitrary (RAPD), semi-arbitrary (ISSR) and sequence based (nuclear rDNA) markers. Trees Struct Funct 32:1083–1101
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Figueira GM, Ramelo PR, Ogasawara DC, Montanari I Jr, Zucchi MI, Cavallari MM, Foglio MA (2010) A set of microsatellite markers for Arrabidaea chica (Bignoniaceae), a medicinal liana from neotropics. Am J Bot 97:e63–e64
Gadissa F, Tesfaye K, Dagne K, Geleta M (2018) Genetic diversity and population structure analyses of Plectranthus edulis (Vatke) Agnew collections from diverse agro-ecologies in Ethiopia using newly developed EST-SSRs marker system. BMC Genet 19:92
Gomez-Fernandez A, Alcocer I, Matesanz S (2016) Does higher connectivity lead to higher genetic diversity? Effects of habitat fragmentation on genetic variation and population structure in a gypsophile. Conserv Genet 17:631–641
Guo W, Wang W, Zhou B, Zhang T (2006) Cross-species transferability of G. arboreum-derived EST-SSRs in the diploid species of Gossypium. Theor Appl Genet 112:1573–1581
Haerinasab M, Rahiminejad MR, Ellison NW (2016) Transferability of simple sequence repeat (SSR) markers developed in red clover (Trifolium pretense L.) to some Trifolium species. Iran J Sci Technol Trans Sci 40:59–62
Harijan Y, Nishanth GK, Katageri IS, Khadi BM (2017) Transferability of heterologous SSR markers to cotton genotypes. Electron J Plant Breed 8:379–384
Heesacker A, Kishore VK, Gao W, Tang S, Kolkman JM, Gingle A, Matvienko M, Kozik A, Michelmore RM, Lai Z, Rieseberg LH, Knapp SJ (2008) SSRs and INDELs mined from the sunflower EST database: abundance, polymorphisms, and cross-taxa utility. Theor Appl Genet 117:1021–1029
Jindal SK, Gupta AK, Kackar NL, Solanki KR (1987a) Variation of quality traits in rohida (Tecomella undulata Sm.) Seem) in situ. In: Khurana DK, Ghosla PK (eds.) IUFRO workshop proceeding. Agroforestry for rural needs vol. II, Indian Society of Tree Scientists, Solan
Jindal SK, Kackar NL, Solanki KR (1987b) Germplasm collection and genetic variability in Rohida (Tecomella undulata (Sm.) Seem) in western Rajasthan. Indian J For 10:52–55
Jones FA, Hubbell SP (2003) Isolation and characterization of microsatellite loci in the tropical tree Jacaranda copaia (Bignoneaceae). Mol Ecol Notes 3:403–405
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177:309–334
Kalia RK, Rai MK, Sharma R, Bhatt RK (2014) Understanding Tecomella undulata: an endangered pharmaceutically important timber species of hot arid regions. Genet Res Crop Evol 61:1397–1421
Kuleung C, Baenziger PS, Dweikat I (2004) Transferability of SSR markers among wheat, rye, and triticale. Theor Appl Genet 108:1147–1150
Lewontin RC (1972) The apportionment of human diversity. Evol Biol 6:391–398
Li YC, Fahima T, Korol AB, Peng J, Roder MS, Kirzhner V, Beiles A, Nevo E (2000) Microsatellite diversity correlated with ecological-edaphic and genetic factors in three microsites of wild emmer wheat in North Israel. Mol Biol Evol 17:851–862
Li X, Li M, Hou L, Zhang Z, Pang X, Li Y (2018) De novo transcriptome assembly and population genetic analysis for an endangered Chinese endemic Acer miaotaiense (Aceraceae). Genes 9:378
Litt M, Luty JA (1989) A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet 44:397–401
Miz RB, Tacuatiá LO, Cidade FW, de Souza AP, Bered F, Eggers L, de Souza-Chies TT (2016) Isolation and characterization of microsatellite loci in Sisyrinchium (Iridaceae) and cross amplification in other genera. Genet Mol Res 15:38474
Mottura MC, Finkeldey R, Verga AR, Gailing O (2005) Development and characterisation of microsatellite markers for Prosopis chilensis and Prosopis flexuosa and cross-amplification. Mol Ecol Note 5:487–489
Negi RS, Sharma MK, Sharma KC, Kshetrapal S, Kothari SL, Trivedi PC (2011) Genetic diversity and variations in the endangered tree (Tecomella undulata) in Rajasthan. Indian J Fundam App Life Sci 1:50–58
Nei M (1973) Analysis of gene diversity in subdivided populations (population structure/genetic variability/heterozygosity/gene differentiation). Proc Natl Acad Sci USA 70(12):3321–3323
Oliveira EJ, Pádua JG, Zucchi MI, Vencovsky R, Vieira MLC (2006) Origin, evolution and genome distribution of microsatellites. Genet Mol Biol 29:294–307
Parida SK, Kalia SK, Sunita K, Dalal V, Hemaprabha G, Selvi A, Pandit A, Singh A, Gaikwad K, Sharma TR, Srivastava PS, Singh NK, Mohapatra T (2009) Informative genomic microsatellite markers for efficient genotyping applications in sugarcane. Theor Appl Genet 118:327–338
Peakall R, Smouse PE (2012) GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pomponio MF, Acuna C, Pentreath V, Lauenstein DL, Poltri SM, Torales S (2015) Characterization of functional SSR markers in Prosopis alba and their transferability across Prosopis species. For Syst 24:04. https://doi.org/10.5424/fs/2015242-07188
Pratik S, Pramod KP, Swagata G, Snehalata M, Nasim A (2016) Confamiliar transferability of simple sequence repeat (SSR) markers from cotton (Gossypium hirsutum L.) and jute (Corchorus olitorius L.) to twenty two Malvaceous species. Biotech 6:1–7
Rai MK, Phulwaria M, Shekhawat NS (2013) Transferability of simple sequence repeat (SSR) markers developed in guava (Psidium guajava L.) to four Myrtaceae species. Mol Biol Rep 40:5067–5071
Rai MK, Shekhawat JK, Kataria V, Shekhawat NS (2017) Cross species transferability and characterization of microsatellite markers in Prosopis cineraria, a multipurpose tree species of Indian Thar Desert. Arid Land Res Manag 31:462–471
Shivakumar MS, Ramesh S, Mohan Rao A, Udaykumar HR, Keerthi CM (2017) Cross legume species/genera transferability of SSR markers and their utility in assessing transferability of SSR markers and their utility in assessing polymorphism among advanced breeding lines in Dolichos bean (Lablab purpureus L.). Int J Curr Microbiol App Sci 6(8):656–668. https://doi.org/10.20546/ijcmas.2017.608.083
Smith JSC, Chin ECL, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Zeigle J (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparison with data from RFLPs and pedigree. Theor Appl Genet 95:163–173
Sork VL (2016) Gene flow and natural selection shape spatial patterns of genes in tree populations: implications for evolutionary processes and applications. Evol Appl 9(1):291–310
Staton M, Best T, Khodwekar S, Owusu S, Xu T, Xu Y, Jennings T, Cronn R, Arumuganathan AK, Coggeshall M, Gailing O, Liang H, Romero-Severson J, Schlarbaum S, Carlson JE (2015) Preliminary genomic characterization of ten hardwood tree species from multiplexed low coverage whole genome sequencing. PLoS ONE 10(12):e0145031
Vinson CC, Sampaio I, Ciampi Y (2008) Eight variable microsatellite loci for a neotropical tree, Jacaranda copaia (Aubl.) D. Don (bignoniaceae). Mol Ecol Resour 8:1288–1290
Whankaew S, Kanjanawattanawong S, Phumichai C, Smith DR, Narangajavana J, Triwitayakor K (2011) Cross-genera transferability of (simple sequence repeat) SSR markers among cassava (Manihot esculenta Crantz), rubber tree (Hevea brasiliensis Muell. Arg.) and physic nut (Jatropha curcas L.). Afr J Biotechnol 10:1768–1776
Yu F, Wang BH, Feng SP, Wang JY, Li WG, Wu YT (2011) Development, characterization, and cross-species/ genera transferability of SSR markers for rubber tree (Hevea brasiliensis). Plant Cell Rep 30:335–344
Acknowledgement
Research work was carried out under DBT, GoI, New Delhi, funded project on Tecomella undulata sanctioned to RKK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by S. Srivastava.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kalia, R.K., Chhajer, S. & Pathak, R. Cross genera transferability of microsatellite markers from other members of family Bignoniaceae to Tecomella undulata (Sm.) Seem. Acta Physiol Plant 42, 151 (2020). https://doi.org/10.1007/s11738-020-03138-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-020-03138-5