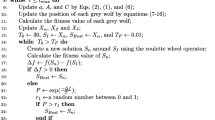Abstract
Breast cancer, as one of the most common diseases threatening the women's life, has attracted serious attention of the clinical and biomedical researchers worldwide. The genome-based studies along with their registered GEO datasets are frequent in the literature. Since several methodologies have been developed for analyzing and identifying gene biomarkers, it is necessary to evaluate their robustness. In this study, three well-known biomarker identification methods (i.e., ClusterOne, MCODE, and BioDiscML) were employed in order to identify the potential biomarkers. Then, the methods were ranked and evaluated using nonlinear classification models developed based on the identified sets of biomarkers. A combined BC microarray dataset consisting of GSE124647, GSE124646, and GSE15852 was used as training set, and two test datasets, GSE15852 and GSE25066, were used for the performance measurement of the trained models. The validation of the proposed models was carried out internally (leave-one-out, fivefold and tenfold cross-validation, random sampling, test on training set) and externally (test on test set). The results showed that ClusterOne, MCODE, and BioDiscML tools ranked first, second, and third, respectively, based on the area under the curve (AUC), accuracy, F1 score, precision, and recall metrics. Overall, it can be concluded that the descriptive values of gene biomarkers in terms of their biological aspects that have been determined by a given methodology and the predictive power of the models developed based on the identified gene biomarkers should be considered simultaneously while validating the biomarker identification approaches.





Similar content being viewed by others
References
Smith RA, Andrews KS, Brooks D, Fedewa SA, Manassaram-Baptiste D, Saslow D, Wender RC (2019) Cancer screening in the United States, 2019: a review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin 69(3):184–210. https://doi.org/10.3322/caac.21557
Malvezzi M, Carioli G, Bertuccio P, Boffetta P, Levi F, La Vecchia C, Negri E (2019) European cancer mortality predictions for the year 2019 with focus on breast cancer. Ann Oncol 30(5):781–787. https://doi.org/10.1093/annonc/mdz051
Khuwaja GA, Abu-Rezq AN (2004) Bimodal breast cancer classification system. Pattern Anal Appl 7(3):235–242. https://doi.org/10.1007/BF02683990
Savage P, Yu N, Dumitra S, Meterissian S (2019) The effect of the American Joint Committee on Cancer eighth edition on breast cancer staging and prognostication. Eur J Surg Oncol 45(10):1817–1820. https://doi.org/10.1016/j.ejso.2019.03.027
Ma X-J, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM (2003) Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci 100(10):5974–5979. https://doi.org/10.1073/pnas.0931261100
Lai J, Wang H, Pan Z, Su F (2019) A novel six-microRNA-based model to improve prognosis prediction of breast cancer. Aging (Albany NY) 11(2):649. https://doi.org/10.18632/aging.101767
Alickovic E, Subasi A (2020) Normalized neural networks for breast cancer classification. In: Badnjevic A, Škrbić R, L GP (eds) CMBEBIH 2019. Springer, Cham, pp 519–524. https://doi.org/10.1007/978-3-030-17971-7_77
Sheth D, Giger ML (2019) Artificial intelligence in the interpretation of breast cancer on MRI. J Magn Reson Imaging. https://doi.org/10.1002/jmri.26878
Rodriguez-Ruiz A, Lång K, Gubern-Merida A, Broeders M, Gennaro G, Clauser P, Helbich TH, Chevalier M, Tan T, Mertelmeier T (2019) Stand-alone artificial intelligence for breast cancer detection in mammography: comparison with 101 radiologists. JNCI J Natl Cancer Inst 111(9):916–922. https://doi.org/10.1093/jnci/djy222
Shamai G, Binenbaum Y, Slossberg R, Duek I, Gil Z, Kimmel R (2019) Artificial intelligence algorithms to assess hormonal status from tissue microarrays in patients with breast cancer. JAMA Netw Open 2(7):e197700–e197700. https://doi.org/10.1001/jamanetworkopen.2019.7700
Wu G-G, Zhou L-Q, Xu J-W, Wang J-Y, Wei Q, Deng Y-B, Cui X-W, Dietrich CF (2019) Artificial intelligence in breast ultrasound. World J Radiol 11(2):19. https://doi.org/10.4329/wjr.v11.i2.19
Le E, Wang Y, Huang Y, Hickman S, Gilbert F (2019) Artificial intelligence in breast imaging. Clin Radiol. https://doi.org/10.1016/j.crad.2019.02.006
Van't Veer LJ, Dai H, Van De Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, Van Der Kooy K, Marton MJ, Witteveen AT (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415(6871):530. https://doi.org/10.1038/415530a
Al-Quraishi T, Abawajy JH, Al-Quraishi N, Abdalrada A, Al-Omairi L (2019) Predicting breast cancer risk using subset of genes. In: 2019 6th international conference on control, decision and information technologies (CoDIT), 23–26 April 2019. pp 1379–1384. https://doi.org/10.1109/CoDIT.2019.8820378
Ferroni P, Zanzotto FM, Riondino S, Scarpato N, Guadagni F, Roselli M (2019) Breast cancer prognosis using a machine learning approach. Cancers 11(3):328. https://doi.org/10.3390/cancers11030328
Liu Q, Hu P (2019) Association analysis of deep genomic features extracted by denoising autoencoders in breast cancer. Cancers 11(4):494. https://doi.org/10.3390/cancers11040494
Almeida PP, Cardoso CP, de Freitas LM (2020) PDAC-ANN: an artificial neural network to predict pancreatic ductal adenocarcinoma based on gene expression. BMC Cancer 20(1):82. https://doi.org/10.1186/s12885-020-6533-0
Amjad E, Asnaashari S, Sokouti B, Dastmalchi S (2020) Systems biology comprehensive analysis on breast cancer for identification of key gene modules and genes associated with TNM-based clinical stages. Sci Rep 10(1):10816. https://doi.org/10.1038/s41598-020-67643-w
Nepusz T, Yu H, Paccanaro A (2012) Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods 9(5):471–472. https://doi.org/10.1038/nmeth.1938
Bader GD, Hogue CWV (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform 4:2–2. https://doi.org/10.1186/1471-2105-4-2
Leclercq M, Vittrant B, Martin-Magniette ML, Scott Boyer MP, Perin O, Bergeron A, Fradet Y, Droit A (2019) Large-scale automatic feature selection for biomarker discovery in high-dimensional OMICs data. Front Genet 10:452–452. https://doi.org/10.3389/fgene.2019.00452
Demšar J, Curk T, Erjavec A, Gorup Č, Hočevar T, Milutinovič M, Možina M, Polajnar M, Toplak M, Starič A, Štajdohar M, Umek L, Žagar L, Žbontar J, Žitnik M, Zupan B (2013) Orange: data mining toolbox in python. J Mach Learn Res 14 (1):2349–2353. https://orange.biolab.si/citation/
Kawakami E, Tabata J, Yanaihara N, Ishikawa T, Koseki K, Iida Y, Saito M, Komazaki H, Shapiro JS, Goto C, Akiyama Y, Saito R, Saito M, Takano H, Yamada K, Okamoto A (2019) Application of artificial intelligence for preoperative diagnostic and prognostic prediction in epithelial ovarian cancer based on blood biomarkers. Clin Cancer Res. https://doi.org/10.1158/1078-0432.CCR-18-3378
Moitra D, Mandal RK (2019) Automated AJCC staging of non-small cell lung cancer (NSCLC) using deep convolutional neural network (CNN) and recurrent neural network (RNN). Health Inf Sci Syst 7(1):14. https://doi.org/10.1007/s13755-019-0077-1
Lin Y, Qian F, Shen L, Chen F, Chen J, Shen B (2019) Computer-aided biomarker discovery for precision medicine: data resources, models and applications. Brief Bioinform 20(3):952–975. https://doi.org/10.1093/bib/bbx158
Qi X, Lin Y, Chen J, Shen B (2020) Decoding competing endogenous RNA networks for cancer biomarker discovery. Brief Bioinform 21(2):441–457. https://doi.org/10.1093/bib/bbz006
Chen J, Sun M, Shen B (2015) Deciphering oncogenic drivers: from single genes to integrated pathways. Brief Bioinform 16(3):413–428. https://doi.org/10.1093/bib/bbu039
Acknowledgements
The authors would like to thank the Research Office of Tabriz University of Medical Sciences for providing financial support scheme (Grant No. 64904).
Funding
The work has been supported by Grant No. 64904.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors confirm that there are no conflicts of interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Amjad, E., Asnaashari, S., Sokouti, B. et al. Impact of Gene Biomarker Discovery Tools Based on Protein–Protein Interaction and Machine Learning on Performance of Artificial Intelligence Models in Predicting Clinical Stages of Breast Cancer. Interdiscip Sci Comput Life Sci 12, 476–486 (2020). https://doi.org/10.1007/s12539-020-00390-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-020-00390-8