Abstract
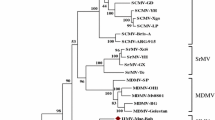
Morphological, biological, serological, and molecular tests underpin the description of costus stripe mosaic virus (CoSMV) as a new member of the genus Potyvirus, family Potyviridae. Found affecting the native ornamental Costus spiralis in Brazil, the pathogen showed a severely restricted natural and experimental host range. Excluding the poly(A) tail, the CoSMV genome contains a large open reading frame (ORF) of 9,446 nucleotides that encodes a polyprotein with 3,046 amino acids, which is potentially cleaved into ten products, and a small ORF (77 amino acids) knows as PIPO. Genome analysis demonstrated the highest CoSMV nucleotide sequence identity to onion yellow dwarf virus (51.79%). No evidence of recombination was detected in the CoSMV genome, and phylogenetic analysis revealed its basal position in a group formed by members of the genus Potyvirus, along with Cyrtanthus elatus virus A (Vallota speciosa virus) and canna yellow streak virus. CoSMV was not transmitted by aphids of the species Aphis solanella, Myzus persicae or Uroleucon sonchi, which could be due to mutations in the HC-Pro motifs required for aphid transmission. A divergence in the P1 protein cleavage site was found when compared to other members of the family Potyviridae. Based on its unique biological and molecular characteristics and the current species demarcation criteria, we propose CoSMV to be a new tentative member of the genus Potyvirus.



Similar content being viewed by others
References
Souza CS, Lorenzi H (2012) Botânica Sistemática. Guia ilustrado para identificação das famílias de Fanerógamas nativas e exóticas no Brasil, baseado em APG III. 3ed. Nova Odessa, SP, Brazil
Myers N, Mittermeier RA, Mittermeier CG, Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Castro CEF, Moreira SR, Castro ACR, Souza FVD, Loges V, Gonçalves C, Costa MAPC, Moura LF (2011) Avaliação de espécies de Costaceae para uso ornamental. Rev Bras Hort Orn 17(1):63–74
Skinner D (2016) Ornamental costus. Orn Hort 22:307–317
Duraipandiyan V, Al-Harbi NA, Ignacimuthu S, Muthukumar C (2012) Antimicrobial activity of sesquiterpene lactones isolated from medicinal plant, Costus speciosus (Koen ex. Retz.) Sm. BioMedCentral Compl Altern Med 12:13. https://doi.org/10.1186/1472-6882-12-13
International Committee on Taxonomy of Viruses (2017) https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/w/potyviridae. Accessed 16 jun 2020
Wylie SJ, Adam M, Chalam C, Kreuze J, López-Moya JJ, Ohshima K, Praveen S, Rabenstein F, Stenger D, Wang A, Zerbini FM (2017) ICTV report consortium, ICTV Virus Taxonomy Profile: Potyviridae. J Gen Virol 98:352–354
Chung BY, Miller WA, Atkins JF, Firth AE (2008) An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci USA 105:5897–5902. https://doi.org/10.1073/pnas.0800468105
Olspert A, Chung BY, Atkins JF, Carr JP, Firth AE (2015) Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep 16:995–1004. https://doi.org/10.15252/embr.201540509
Rodamilans B, Valli A, Mingot A, San León D, Baulcombe D, López-Moya JJ, García JA (2015) RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viroses of the Potyviridae family. J Virol 89(13):6965–6967. https://doi.org/10.1128/JVI00337-15
Kitajima EW, Nome CF (1999) Microscopia electrónica em virologia vegetal. In: Docampo DM, Lenardon SL (eds) Métodos para Detectar Patógenos Sistémicos. IFFIVE/INTA/JICA, Córdoba, pp 59–87
Ha C, Coombs S, Revill PA, Harding RM, Vu M, Dale JL (2008) Design and application of two novel degenerate primer pairs for the detection and complete genomic characterization of potyviruses. Arch Virol 153:25–36
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using Diamond program. Nat Meth 12:59–60
Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9:868–877
Laetsch D, Blaxter M (2017) BlobTools: Interrogation of genome assemblies [version 1; referees: 2 approved with reservations], F1000Research 6
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinform 10:421. https://doi.org/10.1186/1471-2105-10-421
Kumar S, Jones M, Koutsopoulos G, Clarke M, Blaxter M (2013) Blobology: exploring raw genome data for contaminants, symbionts and parasites using taxon-annotated GC-coverage plots. Front Genet 4:237
Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, Raytselis Y, Sayers EW, Tao T, Ye J, Zaretskaya I (2013) BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41:W29–W33. https://doi.org/10.1093/nar/gkt282
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D (2013) Using tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14:193–202. https://doi.org/10.1093/bib/bbs012
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Criscuolo A, Gribaldo S (2020) BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol 10:210. https://doi.org/10.1186/1471-2148-10-210
Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25(7):1307–1320. https://doi.org/10.1093/molbev/msn067
Trifinopoulos J, Nguyen LT, Haeseler A, Minh BQ (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235. https://doi.org/10.1093/nar/gkw256
Letunic I, Bork P (2019) Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47(W1):W256–W259. https://doi.org/10.1093/nar/gkz239
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) Molecular evolutionary genetics analyses across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/myy096
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. https://doi.org/10.1093/VE/VEV003
Adams MJ, Antoniw JF, Beaudoin F (2005) Overview and analysis of the polyprotein cleavage sites in the Family Potyviridae. Mol Plant Pathol 6:471–480
Edwardson JR (1974) Some properties of the potato virus Y-group. Fla Agric Expt Stn Monog Ser. No 5, p 398
Chen J, Shi Y-H, Li M-Y, Adams MJ, Chen J-P (2008) A new potyvirus from butterfly flower (Iris japonica Thunb) in Zhejiang, China. Arch Virol 153:567–569. https://doi.org/10.1007/s00705-007-0014-5
Nigam D, Latourrette K, Souza PFN, Garcia-Ruiz H (2019) Genome-wide variation in Potyviruses. Front Plant Sci 10:1439. https://doi.org/10.3389/fpls.2019.01439
Blackman RL, Eastop VF (2007) Taxonomic issues. In: van Emden VF, Harrington R (eds) Aphids as crop pests. CAB International, Wallingford, pp 1–29
Baulcombe DC, Lloyd J, Manoussopoulus IN, Roberts IM, Harrison BD (1993) Signal for potyvirus-dependent aphid transmission of potato aucuba mosaic virus and the effect of its transfer to potato virus X. J Gen Virol 74:1245–1253. https://doi.org/10.1099/0022-1317-74-7-1245
Kondo H, Maeda T, Gara IW, Chiba S, Maruyama K, Tamada T, Suzuki N (2014) Complete genome sequence of Habenaria mosaic virus, a new potyvirus infecting terrestrial orchids (habenaria radiata) in Japan. Arch Virol 159:163–166. https://doi.org/10.1007/s00705-013-1784-6
Mollov D, Lockhart B, Zlesak D (2013) Complete nucleotide sequence of rose yellow mosaic virus, a novel member of the family Potyviridae. Arch Virol 158:1917–1923
Yuan W, Du K, Zhou T (2017) Complete genomic sequence of common reed chlorotic virus, a novel member of the family Potyviridae. Arch Virol 162:3541–3544. https://doi.org/10.1007/s00705-017-3454-6
Seo J-K, Kim M-K, Kwak H-R, Kim J-S, Choi H-S (2017) Complete genome sequence of longan witches’ broom-associated virus, a novel member of the family Potyviridae. Arch Virol 162:2885–2889. https://doi.org/10.1007/s00705-017-3405-2
Gibbs AJ, Hajizadeh M, Ohshima K, Jones AC (2020) The potyviruses: an evolutionary synthesis is emerging. Viruses 12:132. https://doi.org/10.3390/v12020132
Acknowledgements
We are grateful to Marcela Paduan and Horacio Montenegro (Center for Functional Genomics, ESALQ, USP) for their excellent technical support. We also thank Charleston Gonçalves (Instituto Agronômico de Campinas, IAC, SP) for the botanical identification of Costus species, and IAC Horticulture Center, which provided the Costus species from the Ornamental Zingiberales Collection used in the transmission tests. We thank Juliana Freitas-Astúa (EMBRAPA Cassava and Fruits, BA, Brazil) for providing laboratory facilities for RACE analysis.
Funding
This research received financial support from FAPESP 2017/18910-4 and 50334-3.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Stephen John Wylie.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Alexandre, M.A.V., Duarte, L.M.L., Ramos-González, P.L. et al. Costus stripe mosaic virus, a tentative new member of the genus Potyvirus. Arch Virol 165, 2541–2548 (2020). https://doi.org/10.1007/s00705-020-04788-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04788-z