Abstract
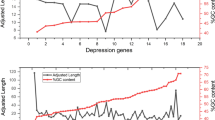
Codon usage bias (CUB) is the unequal usage of synonymous codon; some codons are more preferred than others. CUB analysis has applications in understanding the molecular organization of genome, genetics, gene expression, and molecular evolution. Bioinformatic approach was used to analyze the protein-coding sequences of genes involved in the anxiety to understand the patterns of codon usage as no work was reported yet. The improved effective number of codons (Nc) values ranged from 43.55 to 55.06, with a mean of 44.57, suggested that the overall CUB was low for genes associated with anxiety. The overall GC and AT content was 54.76 and 45.24, respectively. Relative synonymous codon usage (RSCU) analysis revealed that most frequently used codon ended mostly with C or G. The over-represented codons in genes associated with anxiety were CTG, ATC, GTG, AGC, ACC, and GCC, while under-represented codons were TTA, CTT, CTA, ATA, GTT, GTA, TCG, CCG, GCG, CAA, and CGT. Correlation analysis was performed between overall nucleotide composition and its 3rd codon positions, and observed highly significant (p < 0.01) correlation between them suggested that both mutation pressure and natural selection might affect the pattern of CUB. The highly significant correlation (0.598**, p < 0.01) was also observed between GC12 with GC3 suggested that directional mutation pressure might acted on all codon positions for genes associated with anxiety.









Similar content being viewed by others
References
Kjernisted KD, Bleau P (2004) Long-term goals in the management of acute and chronic anxiety disorders. Can J Psychiatr 49:51–63
Weinberger DR (2001) Anxiety at the frontier of molecular medicine. N Engl J Med 344:1247–1249
Clément Y, Calatayud F, Belzung C (2002) Genetic basis of anxiety-like behaviour: a critical review. Brain Res Bull 57:57–71
Gupta V, Bansal P, Kumar S, Sannd R, Rao M (2010) Therapeutic efficacy of phytochemicals as anti-anxiety-a review. J Pharm Res 3:174–179
Jung JW, Yoon BH, Oh HR, Ahn J-H, Kim SY, Park S-Y, Ryu JH (2006) Anxiolytic-like effects of Gastrodia elata and its phenolic constituents in mice. Biol Pharm Bull 29:261–265
Leon AC, Portera L, Weissman MM (1995) The social costs of anxiety disorders. Br J Psychiatry 166:19–22
Beuzen A, Belzung C (1995) Link between emotional memory and anxiety states: a study by principal component analysis. Physiol Behav 58:111–118
Weeks BS (2009) Formulations of dietary supplements and herbal extracts for relaxation and anxiolytic action: Relarian. Med Sci Monit 15:RA256–RA262
Nestoros JN (1984) GABAergic mechanisms and anxiety: an overview and a new neurophysiological model. Can J Psychiatr 29:520–529
Khanum F, Razack S (2010) Anxiety–herbal treatment: a review. Res Rev Biomed Biotech 1:83–89
Barchas JD (1998) Biochemical hypotheses of mood and anxiety disorders. Basic neurochemistry: molecular, cellular and medical aspects (CD-ROM)
Morris-Rosendahl DJ (2002) Are there anxious genes? Dialogues Clin Neurosci 4:251
Lesch K, Greenberg B, Higley J, Bennett A, Murphy D (2002) Serotonin transporter, personality, and behavior: toward dissection of gene–gene and gene–environment interaction. Mol Genet Human Personal:109–136
Bennetzen JL, Hall B (1982) Codon selection in yeast. J Biol Chem 257:3026–3031
Plotkin JB, Robins H, Levine AJ (2004) Tissue-specific codon usage and the expression of human genes. Proc Natl Acad Sci U S A 101:12588–12591
Duret L (2002) Evolution of synonymous codon usage in metazoans. Curr Opin Genet Dev 12:640–649
Biro JC (2008) Studies on the origin and evolution of codon bias. arXiv preprint arXiv:0807.3901
Bulmer M (1991) The selection-mutation-drift theory of synonymous codon usage. Genetics 129:897–907
Ikemura T (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2:13–34
Gupta S, Ghosh T (2001) Gene expressivity is the main factor in dictating the codon usage variation among the genes in Pseudomonas aeruginosa. Gene 273:63–70
Liu Q (2006) Analysis of codon usage pattern in the radioresistant bacterium Deinococcus radiodurans. Biosystems 85:99–106
Yang X, Luo X, Cai X (2014) Analysis of codon usage pattern in Taenia saginata based on a transcriptome dataset. Parasit Vectors 7:1–11
Ahn I, Jeong B-J, Bae S-E, Jung J, Son HS (2006) Genomic analysis of influenza A viruses, including avian flu (H5N1) strains. Eur J Epidemiol 21:511–519
Zheng Y, Zhao W-M, Wang H, Zhou Y-B, Luan Y, Qi M, Cheng Y-Z, Tang W et al (2007) Codon usage bias in Chlamydia trachomatis and the effect of codon modification in the MOMP gene on immune responses to vaccination. This paper is one of a selection of papers in this Special Issue, entitled International Symposium on Recent Advances in Molecular, Clinical, and Social Medicine, and has undergone the Journal's usual peer-review process. Biochem Cell Biol 85:218–226
Kane JF (1995) Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol 6:494–500
Gupta S, Bhattacharyya T, Ghosh TC (2004) Synonymous codon usage in Lactococcus lactis: mutational bias versus translational selection. J Biomol Struct Dyn 21:527–535
Lin K, Kuang Y, Joseph JS, Kolatkar PR (2002) Conserved codon composition of ribosomal protein coding genes in Escherichia coli, Mycobacterium tuberculosis and Saccharomyces cerevisiae: lessons from supervised machine learning in functional genomics. Nucleic Acids Res 30:2599–2607
Erhardt A, Czibere L, Roeske D, Lucae S, Unschuld PG, Ripke S, Specht M, Kohli MA et al (2011) TMEM132D, a new candidate for anxiety phenotypes: evidence from human and mouse studies. Mol Psychiatry 16:647–663
Schinka J, Busch R, Robichaux-Keene N (2004) A meta-analysis of the association between the serotonin transporter gene polymorphism (5-HTTLPR) and trait anxiety. Mol Psychiatry 9:197–202
Sharp PM, Li W-H (1986) An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol 24:28–38
Wright F (1990) The ‘effective number of codons’ used in a gene. Gene 87:23–29
Satapathy SS, Sahoo AK, Ray SK, Ghosh TC (2017) Codon degeneracy and amino acid abundance influence the measures of codon usage bias: improved Nc (N̂c) and ENCprime (N̂′ c) measures. Genes Cells 22:277–283
Grantham R, Gautier C, Gouy M (1980) Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res 8:1893–1912
Shields DC, Sharp PM (1987) Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res 15:8023–8040
Sueoka N (1999) Two aspects of DNA base composition: G+ C content and translation-coupled deviation from intra-strand rule of A= T and G= C. J Mol Evol 49:49–62
Sueoka N (1995) Intrastrand parity rules of DNA base composition and usage biases of synonymous codons. J Mol Evol 40:318–325
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Puigbò P, Bravo IG, Garcia-Vallve S (2008) CAIcal: a combined set of tools to assess codon usage adaptation. Biol Direct 3:1–8
Moura G, Pinheiro M, Arrais J, Gomes AC, Carreto L, Freitas A, Oliveira JL, Santos MA (2007) Large scale comparative codon-pair context analysis unveils general rules that fine-tune evolution of mRNA primary structure. PLoS One 2:e847
Zhang Z, Dai W, Dai D (2013) Synonymous codon usage in TTSuV2: analysis and comparison with TTSuV1. PLoS One 8:e81469
Jenkins GM, Holmes EC (2003) The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res 92:1–7
Behura SK, Severson DW (2012) Comparative analysis of codon usage bias and codon context patterns between dipteran and hymenopteran sequenced genomes. PLoS One 7:e43111
Wei L, He J, Jia X, Qi Q, Liang Z, Zheng H, Ping Y, Liu S et al (2014) Analysis of codon usage bias of mitochondrial genome in Bombyx mori and its relation to evolution. BMC Evol Biol 14:262
Uddin A, Chakraborty S (2016) Codon usage trend in mitochondrial CYB gene. Gene 586:105–114
Sueoka N (1988) Directional mutation pressure and neutral molecular evolution. Proc Natl Acad Sci 85:2653–2657
Zhao Y, Zheng H, Xu A, Yan D, Jiang Z, Qi Q, Sun J (2016) Analysis of codon usage bias of envelope glycoprotein genes in nuclear polyhedrosis virus (NPV) and its relation to evolution. BMC Genomics 17:677
Uddin M, Kabir M, Niaz K, Jeandet P, Clément C, Mathew B, Rauf A, Rengasamy KR et al (2020) Molecular insight into the therapeutic promise of flavonoids against Alzheimer’s disease. Molecules 25:1267
Uddin A, Chakraborty S (2018) Codon usage pattern of genes involved in central nervous system. Mol Neurobiol:1–12
Fuller SJ, Sohrabi HR, Goozee KG, Sankaranarayanan A, Martins RN (2019) Alzheimer's disease and other neurodegenerative diseases. Neurodegener Alzheimer Dis:9–42
Choudhury MN, Uddin A, Chakraborty S (2017) Codon usage bias and its influencing factors for Y-linked genes in human. Comput Biol Chem
Schwartz S, Meshorer E, Ast G (2009) Chromatin organization marks exon-intron structure. Nat Struct Mol Biol 16:990–995
Kaufmann WK, Paules RS (1996) DNA damage and cell cycle checkpoints. FASEB J 10:238–247
Choudhury MN, Chakraborty S (2015) Codon usage pattern in human SPANX genes. Bioinformation 11:454–459
Uddin A, Chakraborty S (2015) Synonymous codon usage pattern in mitochondrial CYB gene in pisces, aves, and mammals. Mitochondrial DNA:1–10
Acknowledgments
The author thanks Moinul Hoque Choudhury Memorial Science College, Algapur, Hailakandi, for providing necessary facilities.
Data Accessibility
All data are available in the manuscript and the supplementary file.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The author declares no conflict of interests in this work.
Ethical Approval
The study is based on DNA sequence–based analysis. Ethical clearance is not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Uddin, A. Compositional Features and Codon Usage Pattern of Genes Associated with Anxiety in Human. Mol Neurobiol 57, 4911–4920 (2020). https://doi.org/10.1007/s12035-020-02068-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-020-02068-0