Abstract
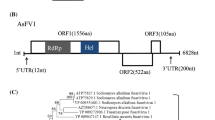
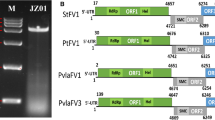
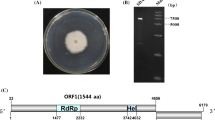
Here, we describe a novel mycovirus, Auricularia heimuer fusarivirus 1 (AhFV1), isolated from the edible fungus Auricularia heimuer strain CCMJ1296. The virus has a single-stranded positive-sense [+ssRNA] genome of 7,127 nucleotides containing two overlapping open reading frames (ORFs) and a poly(A) tail. The large ORF1 encodes a polyprotein of 1,637 amino acids (aa) with conserved RNA-dependent RNA polymerase (RdRp) and DEAD-like helicase superfamily (DEXDc) domains. ORF2 encodes a putative 633-aa protein with unknown function. A BLAST search showed that the nucleotide sequence of the AhFV1 genome is 41.28% identical to that of Sclerotium rolfsii fusarivirus 2 and 40.49% identical to that of Sclerotium rolfsii fusarivirus 1. Phylogenetic analysis based on RdRp and helicase (Hel) sequences indicated that AhFV1 is related to unclassified mycoviruses and other fusariviruses. Our data suggest that AhFV1 should be classified as a member of the newly proposed family “Fusariviridae”. This is the second virus and the first full genome sequence of a fusarivirus from A. heimuer.



Similar content being viewed by others
References
Yaegashi H, Kanematsu S, Ito T (2012) Molecular characterization of a new hypovirus infecting a phytopathogenic, Valsa ceratosperma. Virus Res 165(2):143–150
Wang M, Wang Y, Sun X et al (2015) Characterization of a novel megabirnavirus from Sclerotinia sclerotiorum reveals horizontal gene transfer from ssRNA virus to dsRNA virus. J Virol 89(16):8567–8579
Lin Y, Fujita M, Chiba S et al (2019) Two novel fungal negative-strand RNA viruses related to mymonaviruses and phenuiviruses in the shiitake mushroom (Lentinula edodes). Virology 533:125–136
Revill PA, Wright PJ (1997) RT-PCR detection of dsRNAs associated with La France disease of the cultivated mushroom Agaricus bisporus (Lange) imbach. J Virol Methods 63:17–26
Yu H, Lee J, Lee N et al (2004) Identification of three isometric viruses from Pleurotus ostreatus (Running title: ssRNA and dsRNA viruses of oyster mushroom, (Pleurotus ostreatus). J Huazhong Agric 23(1):150–156
Liu R, Cheng J, Fu Y et al (2015) Molecular characterization of a novel positive-sense, single-stranded RNA virus infecting the plant pathogenic fungus Sclerotinia sclerotiorum. Viruses 7(5):2470–2484
Zhang R, Liu S, Chiba S et al (2014) A novel single-stranded RNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix, with similarity to hypo-like viruses. Front Microbiol 5:360
Liu H, Fu Y, Jiang D et al (2010) Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J Virol 84(22):11876–11887
Grogan HM, Adie BAT, Gaze RH et al (2003) Double-stranded RNA elements associated with the MVX disease of Agaricus bisporus. Mycol Res 107(2):147–154
Ghabrial SA, Castón JR, Jiang D et al (2015) 50-plus years of fungal viruses. Virology 479:356–368
Yu X, Li B, Fu Y et al (2010) A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci 107(18):8387–8392
Yu X, Li B, Fu Y et al (2013) Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc Natl Acad Sci 110(4):1452–1457
Liu L, Xie J, Cheng J et al (2014) Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci USA 111(33):12205–12210
Wang L, He H, Wang S et al (2018) Evidence for a novel negative-stranded RNA mycovirus isolated from the plant pathogenic fungus Fusarium graminearum. Virology 518:232–240
Li K, Zheng D, Cheng J et al (2016) Characterization of a novel Sclerotinia sclerotiorum RNA virus as the prototype of a new proposed family within the order Tymovirales. Virus Res 219:92–99
Hamid M, Xie J, Wu S et al (2018) A novel deltaflexivirus that infects the plant fungal pathogen, Sclerotinia sclerotiorum, can be transmitted among host vegetative incompatible strains. Viruse 10(6):295
Liu W, Hai D, Mu F et al (2020) Molecular characterization of a novel fusarivirus infecting the plant-pathogenic fungus Botryosphaeria dothidea. Adv Virol 165(4):1033–1037
Wu Q, Tan Z, Liu H et al (2010) Chemical characterization of Auricularia auricula polysaccharides and its pharmacological effect on heart antioxidant enzyme activities and left ventricular function in aged mice. Int J Biol Macromol 46(3):284–288
Nguyen L, Wang D, Hu D et al (2012) Immuno-enhancing activity of sulfated Auricularia auricula polysaccharides. Carbohyd Polym 89(4):1117–1122
Yuan Y, Wu F, Si J et al (2017) Whole genome sequence of Auricularia heimuer, (Basidiomycota, Fungi), the third most important cultivated mushroom worldwide. Genomics 2017:S0888754317301830
Hollings M (1962) Viruses associated with a die-back disease of cultivated mushroom. Nature 196:962–965
Elibuyuk IO, Bostan H (2010) Detection of a virus disease on white button mushroom (Agaricus bisporus) in Ankara, Turkey. Int J Agric Biol 12:597–600
Magae Y, Sunagawa M (2010) Characterization of a mycovirus associated with the brown discoloration of edible mushroom, Flammulina velutipes. Virol J 7(1):342
Magae Y, Hayashi N (1999) Double-stranded RNA and virus-like particles in the edible basidiomycete Flammulina velutipes (Enokitake). FEMS Microbiol Lett 180:331–335
Won HK, Park SJ, Kim DK et al (2013) Isolation and characterization of a mycovirus in Lentinula edodes. J Microbiol 51(1):118–122
Barroso G, Labarère J (1990) Evidence for viral and naked double-stranded RNAs in the basidiomycete Agrocybe aegerita. Curr Genet 18(3):231–237
Chang Y, Chen J, Chang K et al (2018) Cloning and expression of the lectin gene from the mushroom Agrocybe aegerita and the activities of recombinant lectin in the resistance of shrimp white spot syndrome virus infection. Dev Comp Immunol 90:1–9
Ro HS, Lee NJ, Lee CW et al (2006) Isolation of novel mycovirus OMIV in Pleurotus ostreatus and its detection using a triple antibody sandwich-ELISA. J Virol Methods 138:24–29
Yu HJ, Lim D, Lee HS (2003) Characterization of a novel single-stranded RNA mycovirus in Pleurotus ostreatus. Virology 314(1):9–15
Li X, Xie J, Hai D et al (2020) Molecular characteristics of a novel ssRNA virus isolated from Auricularia heimuer in China. Adv Virol 165(6):1495–1499
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 30(15):2114–2120
Yuan Y, Wu F, Si J et al (2017) Whole genome sequence of Auricularia heimuer (Basidiomycota, Fungi), the third most important cultivated mushroom worldwide. Genomics 2017:S0888754317301830
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760
Grabherr MG, Haas BJ, Yassour M et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644–652
Darissa O, Willingmann P, Günter A (2010) Optimized approaches for the sequence determination of double-stranded RNA templates. J Virol Methods 169(2):397–403
Lambden PR, Cooke SJ, Caul EO et al (1992) Cloning of noncultivatable human rotavirus by single primer amplification. J Virol 66(3):1817–1822
Kazutaka K, Standley DM (2016) A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 13:1933–1942
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Guindon S, Dufayard JF, Lefort V et al (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59(3):307–321
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding
The National Key Research and Development Program of China (2018YFD1001000, 2017YFD0601002), the Special Fund for Agro-Scientific Research in the Public Interest (201503137), the Jilin Provincial Department of Education (JJKH20180670KJ), and the Program of Introducing Talents of Discipline to Universities (D17014) supported this study.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Ethical approval
This article does not contain any studies involving human participants or animals.
Additional information
Handling Editor: Ioly Kotta-Loizou.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2020_4781_MOESM1_ESM.png
Fig. S1 Comparison of the conserved RdRp amino acid motif of AhFV1 with those of representative viruses of the proposed family “Fusariviridae”
705_2020_4781_MOESM2_ESM.png
Fig. S2 Comparison of the conserved Hel amino acid motif of AhFV1 with those of representative viruses of the proposed family “Fusariviridae”
Rights and permissions
About this article
Cite this article
Li, X., Sui, K., Xie, J. et al. Molecular characterization of a novel fusarivirus infecting the edible fungus Auricularia heimuer. Arch Virol 165, 2689–2693 (2020). https://doi.org/10.1007/s00705-020-04781-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04781-6