Abstract
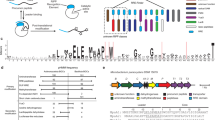
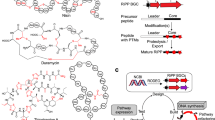
Lasso peptides are a diverse class of ribosomally synthesized and post-translationally modified peptides (RiPPs). Their proteolytic and thermal stability alongside their growing potential as therapeutics has increased attention to these antimicrobial peptides. With the advent of genome mining, the discovery of RiPPs allows for the accurate prediction of putatively encoded structures, however, MSn experiments only provide partial sequence confirmation, therefore 2D NMR experiments are necessary for characterisation. Multiple MS/MS techniques were applied to two structurally characterized lasso peptides, huascopeptin and leepeptin, and one uncharacterized lasso peptide, citrulassin C, which was not isolable in sufficient quantity for NMR analysis. We have shown that MS2 can be used to elucidate the full amino acid sequences previously predicted with genome mining for this compound class. HCD was able to open the macrocycles and fragment the newly opened linear peptides, confirming the complete amino acid sequences of the characterised lasso peptides. In addition, to determine if this technique could be applied at the earliest stages of the isolation process, we targeted a lasso peptide found by genome mining, citrulassin C, and were able to fully elucidate the amino acid sequence using only MS2 from a semi-crude extract of Streptomyces huasconensis HST28T.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hegemann JD, Zimmermann M, Xie X, Marahiel MA. Lasso peptides: an intriguing class of bacterial natural products. Acc Chem Res. 2015;48:1909–19.
Knappe TA, Linne U, Xie X, Marahiel MA. The glucagon receptor antagonist BI-32169 constitutes a new class of lasso peptides. FEBS Lett. 2010;584:785–9.
Iwatsuki M, Tomoda H, Uchida R, Gouda H, Hirono S, Omura S. Lariatins, antimycobacterial peptides produced by Rhodococcus sp. K01-B0171, have a lasso structure. J Am Chem Soc. 2006;128:7486–91.
Detlefsen DJ, Hill SE, Volk KJ, Klohr SE, Tsunakawa M, Furumai T, et al. Siamycins I and II, new anti-HIV-1 peptides: II. Sequence analysis and structure determination of siamycin I. J Antibiot. 1995;48:1515–7.
Gavrish E, Sit CS, Cao S, Kandror O, Spoering A, Peoples A, et al. Lassomycin, a ribosomally synthesized cyclic peptide, kills mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem Biol. 2014;21:509–18.
Metelev M, Tietz JI, Melby JO, Blair PM, Zhu L, Livnat I, et al. Structure, bioactivity, and resistance mechanism of streptomonomicin, an unusual lasso Peptide from an understudied halophilic actinomycete. Chem Biol. 2015;22:241–50.
Um S, Kim YJ, Kwon H, Wen H, Kim SH, Kwon HC, et al. Sungsanpin, a lasso peptide from a deep-sea streptomycete. J Nat Prod. 2013;76:873–9.
Elsayed SS, Trusch F, Deng H, Raab A, Prokes I, Busarakam K, et al. Chaxapeptin, a Lasso Peptide from Extremotolerant Streptomyces leeuwenhoekii Strain C58 from the Hyperarid Atacama Desert. J Org Chem. 2015;80:10252–60.
Weber W, Fischli W, Hochuli E, Kupfer E, Weibel EK. Anantin-a peptide antagonist of the atrial natriuretic factor (ANF). I. Producing organism, fermentation, isolation and biological activity. J Antibiot. 1991;44:164–71.
Potterat O, Wagner K, Gemmecker G, Mack J, Puder C, Vettermann R, et al. BI-32169, a bicyclic 19-peptide with strong glucagon receptor antagonist activity from Streptomyces sp. J Nat Prod. 2004;67:1528–31.
Cheung-Lee WL, Parry ME, Jaramillo Cartagena A, Darst SA, Link AJ. Discovery and structure of the antimicrobial lasso peptide citrocin. J Biol Chem. 2019;294:6822–30.
Kaweewan I, Hemmi H, Komaki H, Harada S, Kodani S. Isolation and structure determination of a new lasso peptide specialicin based on genome mining. Bioorg Medicinal Chem. 2018;26:6050–5.
Santos-Aberturas J, Chandra G, Frattaruolo L, Lacret R, Pham TH, Vior NM, et al. Uncovering the unexplored diversity of thioamidated ribosomal peptides in Actinobacteria using the RiPPER genome mining tool. Nucleic Acids Res. 2019;47:4624–37.
Hudson G, Burkhart B, DiCaprio A, Schwalen C, Kille B, Pogorelov T, et al. Bioinformatic mapping of radical S-adenosylmethionine-dependent ribosomally synthesized and post-translationally modified peptides identifies new Cα, Cβ, and Cγ-linked thioether-containing peptides. J Am Chem Soc. 2019;141:8228–38.
Tietz JI, Schwalen CJ, Patel PS, Maxson T, Blair PM, Tai HC, et al. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat Chem Biol. 2017;13:470–8.
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–7.
Paizs B, Suhai S. Fragmentation pathways of protonated peptides. Mass Spectrom Rev. 2005;24:508–48.
Ngoka LC, Gross ML. Multistep tandem mass spectrometry for sequencing cyclic peptides in an ion-trap mass spectrometer. J Am Soc Mass Spectrom. 1999;10:732–46.
Medzihradszky KF, Chalkley RJ. Lessons in de novo peptide sequencing by tandem mass spectrometry. Mass Spec Rev. 2013;34:43–63.
Mohimani H, Gurevich A, Mikheenko A, Garg N, Nothias L, Ninomiya A, et al. Dereplication of peptidic natural products through database search of mass spectra. Nat Chem Biol. 2017;13:30–7.
Mohimani H, Kersten RD, Liu WT, Wang M, Purvine SO, Wu S, et al. Automated genome mining of ribosomal peptide natural products. ACS Chem Biol. 2014;9:1545–51.
Mohimani H, Yang YL, Liu WT, Hsieh PW, Dorrestein PC, Pevzner PA. Sequencing cyclic peptides by multistage mass spectrometry. Proteomics. 2011;11:3642–50.
Niedermeyer, Timo H J, Strohalm M. mMass as a software tool for the annotation of cyclic peptide tandem mass spectra. PLoS ONE. 2012;7:e44913.
Xie X, Marahiel MA. NMR as an effective tool for the structure determination of lasso peptides. Chem Biochem. 2012;13:621–5.
Rosengren KJ, Clark RJ, Daly NL, Göransson U, Jones A, Craik DJ. Microcin J25 Has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone. J Am Chem Soc. 2003;125:12464–74.
Wilson K, Kalkum M, Ottesen J, Yuzenkova J, Chait BT, Landick R, et al. Structure of microcin J25, a peptide inhibitor of bacterial RNA polymerase, is a lassoed tail. J Am Chem Soc. 2003;125:12475–83.
Jeanne Dit Fouque K, Lavanant H, Zirah S, Hegemann JD, Fage CD, Marahiel MA, et al. General rules of fragmentation evidencing lasso structures in CID and ETD. Analyst. 2018;143:1157–70.
Dit Fouque KJ, Moreno J, Hegemann JD, Zirah S, Rebuffat S, Fernandez-Lima F. Identification of lasso peptide topologies using native nanoelectrospray ionization-trapped ion mobility spectrometry—mass spectrometry. Anal Chem. 2018;90:5139–46.
Cortés-Albayay C, Jarmusch S, Trusch F, Ebel R, Andrews B, Jaspars M, et al. Downsizing class II lasso peptides: genome mining-guided isolation of huascopeptin containing the first Gly1-Asp7 macrocycle. J Org Chem. 2020;85:1661–7.
Gomez-Escribano JP, Castro JF, Razmilic V, Jarmusch SA, Saalbach G, Ebel R, et al. Heterologous expression of a cryptic gene cluster from Streptomyces leeuwenhoekii C34T yields a novel lasso peptide, leepeptin. Appl Environ Microbiol. 2019;85:e01752–19.
Elashal HE, Cohen RD, Elashal HE, Zong C, Link AJ, Raj M. Cyclic and lasso peptides: sequence determination, topology analysis, and rotaxane formation. Angew Chem Int Ed Engl. 2018;57:6150–4.
Brodbelt JS. Ion activation methods for peptides and proteins. Anal Chem. 2016;88:30–51.
Chekan JR, Koos JD, Zong C, Maksimov MO, Link AJ, Nair SK. Structure of the lasso peptide isopeptidase identifies a topology for processing threaded substrates. J Am Chem Soc. 2016;138:16452–8.
Okoro CK, Brown R, Jones AL, Andrews BA, Asenjo JA, Goodfellow M, et al. Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie Van Leeuwenhoek. 2009;95:121–33.
Cortés-Albayay C, Dorador C, Schumann P, Andrews B, Asenjo J, Nouioui I. Streptomyces huasconensis sp. nov., an haloalkalitolerant actinobacterium isolated from a high altitude saline wetland at the Chilean Altiplano. Int J Syst Evolut Microbiol. 2019;69:2315–22.
Rateb ME, Houssen WE, Harrison WT, Deng H, Okoro CK, Asenjo JA, et al. Diverse metabolic profiles of a Streptomyces strain isolated from a hyper-arid environment. J Nat Prod. 2011;74:1965–71.
Goodfellow M, Nouioui I, Sanderson R, Xie F, Bull AT. Rare taxa and dark microbial matter: novel bioactive actinobacteria abound in Atacama Desert soils. Antonie Van Leeuwenhoek. 2018;111:1315–32.
Manfio GP, Atalan E, Zakrzewska-Czerwinska J, Mordarski M, Rodriguez C, Collins MD, et al. Classification of novel soil streptomycetes as Streptomyces aureus sp. nov., Streptomyces laceyi sp. nov. and Streptomyces sanglieri sp. nov. Antonie Van Leeuwenhoek. 2003;83:245–55.
Rateb ME, Houssen WE, Arnold M, Abdelrahman MH, Deng H, Harrison WT, et al. Chaxamycins A-D, bioactive ansamycins from a hyper-arid desert Streptomyces sp. J Nat Prod. 2011;74:1491–9.
Acknowledgements
SAJ would like to thank the University of Aberdeen for an Elphinstone Scholarship. CC-A thanks CONICYT PFCHA/DOCTORADO BECAS CHILE/2016 (#21160585) fellowship and CONICYT Basal Centre Grant for the Centre for Biotechnology and Bioengineering, CeBiB (FB0001). JFC also thanks CONICYT for a National PhD Scholarship (#21110356) and a Visiting Student Scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
This study is dedicated to Professor William Fenical in recognition of his work on establishing marine microbial natural products as a force to be reckoned with.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Jarmusch, S.A., Feldmann, I., Blank-Landeshammer, B. et al. Cutting the Gordian knot: early and complete amino acid sequence confirmation of class II lasso peptides by HCD fragmentation. J Antibiot 73, 772–779 (2020). https://doi.org/10.1038/s41429-020-00369-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-020-00369-z