Abstract
Centrosomes catalyse the formation of microtubules needed to assemble the mitotic spindle apparatus1. Centrosomes themselves duplicate once per cell cycle, in a process that is controlled by the serine/threonine protein kinase PLK4 (refs. 2,3). When PLK4 is chemically inhibited, cell division proceeds without centrosome duplication, generating centrosome-less cells that exhibit delayed, acentrosomal spindle assembly4. Whether PLK4 inhibitors can be leveraged as a treatment for cancer is not yet clear. Here we show that acentrosomal spindle assembly following PLK4 inhibition depends on levels of the centrosomal ubiquitin ligase TRIM37. Low TRIM37 levels accelerate acentrosomal spindle assembly and improve proliferation following PLK4 inhibition, whereas high TRIM37 levels inhibit acentrosomal spindle assembly, leading to mitotic failure and cessation of proliferation. The Chr17q region containing the TRIM37 gene is frequently amplified in neuroblastoma and in breast cancer5,6,7,8, rendering these cancer types highly sensitive to PLK4 inhibition. We find that inactivating TRIM37 improves acentrosomal mitosis because TRIM37 prevents PLK4 from self-assembling into centrosome-independent condensates that serve as ectopic microtubule-organizing centres. By contrast, elevated TRIM37 expression inhibits acentrosomal spindle assembly through a distinct mechanism that involves degradation of the centrosomal component CEP192. Thus, TRIM37 is an essential determinant of mitotic vulnerability to PLK4 inhibition. Linkage of TRIM37 to prevalent cancer-associated genomic changes—including 17q gain in neuroblastoma and 17q23 amplification in breast cancer—may offer an opportunity to use PLK4 inhibition to trigger selective mitotic failure and provide new avenues to treatments for these cancers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The RNA-sequencing data in Extended Data Figs. 4f, g, 5g have been deposited in the National Center for Biotechnology Information (NCBI)’s Gene Expression Omnibus (GEO)42 and can be accessed with GEO accession number GSE148263 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148263). Other data or materials are available from the corresponding authors upon reasonable request. Source data are provided with this paper.
References
Petry, S. Mechanisms of mitotic spindle assembly. Annu. Rev. Biochem. 85, 659–683 (2016).
Nigg, E. A. & Holland, A. J. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 19, 297–312 (2018).
Zitouni, S., Nabais, C., Jana, S. C., Guerrero, A. & Bettencourt-Dias, M. Polo-like kinases: structural variations lead to multiple functions. Nat. Rev. Mol. Cell Biol. 15, 433–452 (2014).
Wong, Y. L. et al. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 348, 1155–1160 (2015).
Bulavin, D. V. et al. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat. Genet. 31, 210–215 (2002).
Ho, N. et al. Delineation of the frequency and boundary of chromosomal copy number variations in paediatric neuroblastoma. Cell Cycle 17, 749–758 (2018).
Li, J. et al. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat. Genet. 31, 133–134 (2002).
Liu, Y. et al. Targeting 17q23 amplicon to overcome the resistance to anti-HER2 therapy in HER2+ breast cancer. Nat. Commun. 9, 4718 (2018).
Woodruff, J. B., Wueseke, O. & Hyman, A. A. Pericentriolar material structure and dynamics. Phil. Trans. R. Soc. Lond. B 369, 20130459 (2014).
Oegema, K., Davis, R. L., Lara-Gonzalez, P., Desai, A. & Shiau, A. K. CFI-400945 is not a selective cellular PLK4 inhibitor. Proc. Natl Acad. Sci. USA 115, E10808–E10809 (2018).
Bazzi, H. & Anderson, K. V. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc. Natl Acad. Sci. USA 111, E1491–E1500 (2014).
Khodjakov, A. & Rieder, C. L. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153, 237–242 (2001).
Meitinger, F. et al. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J. Cell Biol. 214, 155–166 (2016).
Sir, J. H. et al. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J. Cell Biol. 203, 747–756 (2013).
Ma, X. et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555, 371–376 (2018).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Montenegro Gouveia, S. et al. PLK4 is a microtubule-associated protein that self-assembles promoting de novo MTOC formation. J. Cell Sci. 132, jcs219501 (2019).
Park, J. E. et al. Phase separation of Polo-like kinase 4 by autoactivation and clustering drives centriole biogenesis. Nat. Commun. 10, 4959 (2019).
Yamamoto, S. & Kitagawa, D. Self-organization of Plk4 regulates symmetry breaking in centriole duplication. Nat. Commun. 10, 1810 (2019).
Brigant, B., Metzinger-Le Meuth, V., Rochette, J. & Metzinger, L. TRIMming down to TRIM37: relevance to inflammation, cardiovascular disorders, and cancer in MULIBREY nanism. Int. J. Mol. Sci. 20, 67 (2018).
Park, H. H. Structure of TRAF family: current understanding of receptor recognition. Front. Immunol. 9, 1999 (2018).
Bhatnagar, S. et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature 516, 116–120 (2014).
Hu, M. et al. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 4, e27 (2006).
Sheng, Y. et al. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat. Struct. Mol. Biol. 13, 285–291 (2006).
Kim, T. S. et al. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl Acad. Sci. USA 110, E4849–E4857 (2013).
Park, S. Y. et al. Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis. Nat. Struct. Mol. Biol. 21, 696–703 (2014).
Sonnen, K. F., Gabryjonczyk, A. M., Anselm, E., Stierhof, Y. D. & Nigg, E. A. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 126, 3223–3233 (2013).
Joukov, V. & De Nicolo, A. Aurora-PLK1 cascades as key signaling modules in the regulation of mitosis. Sci. Signal. 11, eaar4195 (2018).
Yeow, Z. Y. et al. Targeting TRIM37-driven centrosome dysfunction in 17q23-amplified breast cancer. Nature https://doi.org/10.1038/s41586-020-2690-1 (2020).
Cheung, N. K. & Dyer, M. A. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat. Rev. Cancer 13, 397–411 (2013).
Louis, C. U. & Shohet, J. M. Neuroblastoma: molecular pathogenesis and therapy. Annu. Rev. Med. 66, 49–63 (2015).
Maris, J. M. Recent advances in neuroblastoma. N. Engl. J. Med. 362, 2202–2211 (2010).
Oeffinger, K. C. et al. Chronic health conditions in adult survivors of childhood cancer. N. Engl. J. Med. 355, 1572–1582 (2006).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protocols 8, 2281–2308 (2013).
Brinkman, E. K., Chen, T., Amendola, M. & van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
Kaulich, M. & Dowdy, S. F. Combining CRISPR/Cas9 and rAAV templates for efficient gene editing. Nucleic Acid Ther. 25, 287–296 (2015).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Kamitani, T., Kito, K., Nguyen, H. P. & Yeh, E. T. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem. 272, 28557–28562 (1997).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Heberle, H., Meirelles, G. V., da Silva, F. R., Telles, G. P. & Minghim, R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16, 169 (2015).
Edgar, R., Domrachev, M. & Lash, A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Bauer, M., Cubizolles, F., Schmidt, A. & Nigg, E. A. Quantitative analysis of human centrosome architecture by targeted proteomics and fluorescence imaging. EMBO J. 35, 2152–2166 (2016).
Acknowledgements
We dedicate this manuscript to the memory of T. Gahman, our colleague and friend, with whom we developed centrinone. This work was supported by a National Institutes of Health (NIH) grant to K.O. (grant GM074207) and by funds from the Hilton Ludwig Cancer Prevention Initiative to A.K.S. F.M. was supported by the German Science Foundation (grant ME 4713/1-1). M.O. and S.W. were supported by the Japan Society for the Promotion of Science. R.L.D, J.V.A, D.A.J., A.K.S., A.D. and K.O. received salary and other support from the Ludwig Institute for Cancer Research. We thank P. Zage for the KPNYN cells.
Author information
Authors and Affiliations
Contributions
F.M., A.D. and K.O. conceived and designed the study and wrote the manuscript with the support of A.K.S.; F.M. performed all experiments and analysed data unless otherwise noted; M.O. and K.-Y.L. performed all of the coexpression and interaction analyses; R.L.D. and A.K.S. made the initial observation of the sensitivity of neuroblastoma cell lines to centrinone and helped to design and execute tumour xenograft experiments; A.K.S. designed the TRIM37 TRAF-domain mutant; S.W. analysed the coalescence of pericentriolar material; J.V.A. and R.K. helped to analyse the proliferation of the neuroblastoma cell line; D.A.J. helped to analyse gene copy numbers; M.O., K-Y.L., S.W. and R.L.D. contributed to editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
K.O., A.K.S., A.D., F.M. and R.L.D. are inventors on a pending patent application from the Ludwig Institute for Cancer Research, PCT/US2018/064243: ‘Methods of treating cancer with PLK4 inhibitors’. A.K.S. is also an inventor on a patent application from the Ludwig Institute for Cancer Research, application number PCT/IB2016/000594: ‘Plk4 inhibitors’.
Additional information
Peer review information Nature thanks Renata Basto, Sarah McClelland and William Weiss for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Effect of varying TRIM37 levels on sensitivity to PLK4 inhibition, and TRIM37 expression profile in patient-derived tumours and cancer cell lines.
a, Unique cell line identifiers are used to describe the cell lines in each experiment. The RCL prefix refers to cell lines received from an external source, such as the ATCC. The ODCL prefix refers to cell lines engineered in the Oegema and Desai labs (OD) from received cell lines. b, Top, cell-line code and bottom, experimental protocol for the analysis of mitotic duration and chromosome-segregation failure. Clone 5 is the Tet-ON–TRIM37 cell line shown in Fig. 1a, b, which overexpresses TRIM37 roughly fourfold relative to parental RPE1 cells. c, Immunoblots of the RPE1 cell lines described in b; transgene-encoded TetON–TRIM37 expression was induced for 24 h. α-tub, α-tubulin. d, e, Graphs plotting mitotic duration (d) and the frequency of chromosome-segregation failure (e) following treatment with DMSO (−) versus centrinone (+) for the three analysed cell lines shown in Fig. 1a, b; n = 50 for each condition. Error bars represent 95% confidence intervals. f, Live-imaging-based analysis was used to measure mitotic duration and segregation failure for the cell lines described in b; values are plotted versus TRIM37 protein level measured by semiquantitative western blotting. Each cell line was treated with DMSO (grey) or centrinone (red) and doxycycline before live imaging; the experimental scheme is shown in b. Fifty cells were analysed per condition. Error bars represent 95% confidence intervals. In DMSO, the analysed cell lines exhibited normal mitotic duration and segregation fidelity regardless of TRIM37 protein level. By contrast, in centrinone, loss of TRIM37 reduced mitotic duration and the percentage of cells experiencing segregation failure (green shading), whereas increased TRIM37 protein levels led to a proportional increase in mitotic duration and segregation-failure rate (red shading). g, Left, graph plotting TRIM37 mRNA levels in 2,120 paediatric tumours representing 13 different cancer types (data are from the St Jude PeCan Data Portal15). All paediatric cancer types with more than ten tumours analysed are shown. Values for individual tumours (dots) and median values (black lines) are plotted. The three-letter codes to the right of the graph describe the 13 paediatric cancer types. Neuroblastoma (NBL) tumours exhibit the highest TRIM37 expression. Right, box-and-whiskers plot, comparing TRIM37 mRNA levels in neuroblastoma tumours to those in all other paediatric cancer type tumours. The range represents the 10th to 90th percentiles of the data; the P-value shown is from an unpaired t-test. ****P < 0.0001. FPKM, fragments per kilobase of transcript per million mapped reads. h, Graph plotting TRIM37 mRNA levels across cancer cell lines described in the Cancer Cell Line Encyclopedia (CCLE16; https://portals.broadinstitute.org/ccle). i, mRNA expression versus copy number from CCLE data for breast cancer cell lines. Two cell lines with high TRIM37 copy number and expression (MCF7 and BT474; red), as well as two cell lines with normal copy number and expression (MDA-MB-231 and BT549; green) are marked. j, List of breast cancer and neuroblastoma cell lines used for analysis in Fig. 1d, e. HepG2 is a hepatocellular carcinoma derived cell line with similar TRIM37 expression to control RPE1 cells. For gel source data see Supplementary Fig. 1. For details on statistics, see Methods; unpaired t-tests assuming equal standard deviation were performed.
Extended Data Fig. 2 Analysis of TRIM37 protein levels and centrinone efficacy in different cancer cell lines, and comparison of mitosis in RPE1 and CHP134 neuroblastoma cells following centrinone treatment.
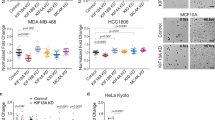
a, Immunoblots used to quantify TRIM37 protein levels across different cell lines. TRIM37 immunoblots are shown above the corresponding Ponceau-stained blots. MCF7 cells have the highest TRIM37 transcript levels and copy number in the CCLE (Extended Data Fig. 1i). Serial dilutions of MCF7 extracts were loaded next to extracts from other cell lines on each blot, and TRIM37 band intensities across a serial dilution of MCF7 cell extract were used to generate a standard curve (graphs below each blot); measured intensities for other cell line extracts were converted into relative expression values using the standard curve. TRIM37 protein level in HepG2 cells was set to 1 and values measured for other cell lines were plotted relative to the HepG2 level in Fig. 1d. b, Protocols used to measure TRIM37 protein levels (left) and conduct passaging-based proliferation analysis of cancer cell lines (right). c, Comparison of TRIM37 mRNA and protein levels across cancer cell lines. mRNA levels are from the CCLE and were transformed from a logarithmic (base 2) to a linear scale. Protein levels are mean values from two measurements, taken as in a, and are plotted relative to the amount of TRIM37 in HepG2, a non-amplified cancer cell line. The inset graph excludes MCF7, which shows exceptionally high TRIM37 mRNA and protein levels. d, Quantification of centrosome number in the indicated cell lines and conditions (n = 100 for each condition). Centrosomes were defined as co-localized foci of CEP192 and γ-tubulin in fixed interphase cells. In the absence of any treatments, there is mild centrosome amplification in the breast cancer cell lines and in one neuroblastoma cell line. Following an eight-day treatment with centrinone, a substantial proportion of the cells from cell lines with relatively low sensitivity to centrinone lacked centrosomes. e, Method used to generate a pool of cells expressing H2b–mRFP for the indicated cell lines. TRIM37 protein levels are shown relative to levels in RPE1 cells, measured by semiquantitative immunoblotting. f, Images are stills from time-lapse sequences of H2b–RFP-expressing mitotic RPE1 and CHP134 cells. Both cell lines exhibit rapid mitosis (taking around 30 min) with no segregation failure in DMSO. Following centrinone treatment, CHP134 cells exhibit more delayed mitosis and higher rates of segregation failure compared with RPE1 cells. Scale bar, 10 μm. g, Quantification of mitotic duration and segregation failure, comparing RPE1 and CHP134 cells. h, Protocols used to analyse mitotic duration, segregation failure and viability of the CHP134-derived cell lines with different levels of TRIM37 protein. For details on statistics, see Methods; unpaired t-tests assuming equal standard deviation were performed.
Extended Data Fig. 3 Analysis of TRIM37∆ cells, rescue with TRIM37 variants and generation of the inducible PLK4 knockout.
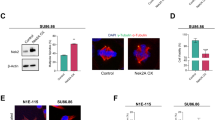
a, Formation of PLK4 condensates in CHP134 neuroblastoma cells with reduced TRIM37 expression. Parental CHP134 cells, which have four copies of the TRIM37 gene, were compared with clone 1 (Fig. 1f; roughly 12% TRIM37 expression relative to parental CHP134). PLK4 condensates were observed in 23% of the clone 1 cells with reduced TRIM37 expression but in none of the parental cells (n = 100 for each). b, Immunoblots of RPE1 cells, comparing the effect on PLK4 protein levels of TRIM37 deletion (top) versus inhibition of PLK4 kinase activity using centrinone (bottom). PLK4 protein levels were elevated roughly sevenfold following the inhibition of kinase activity (7.4 ± 1.1 fold; mean ± s.d.; n = 3), confirming that the detected band corresponds to PLK4. The TRIM37∆ blot is the same as in Fig. 2b. c, Immunofluorescence images of the indicated centrosomal components in TRIM37∆ cells. Scale bar, 10 μm. d, Summary of immunofluorescence analysis in interphase cells. PCM, pericentriolar material. e, Immunofluorescence image showing microtubule organization by a PLK4 condensate in a mitotic TRIM37∆ cell. Scale bar, 10 μm. f, Top, protocol used to conduct live imaging of CEP192 and microtubules. Bottom, images of control and centrinone-treated TRIM37∆ cells with in situ mNG-tagged CEP192 and a transgene that expresses a red fluorescent microtubule-binding domain (MBD). Times in minutes after NEBD are noted on each panel. Scale bar, 10 μm. The merged TRIM37∆ images are the same as those shown in Fig. 2e. g, Description and validation of the inducible PLK4 knockout engineered in TRIM37∆ and control (USP28∆) cells. USP28∆ cells were used as the control because inactivation of USP28 prevents the p53 activation and G1 arrest that are observed as a consequence of delayed mitosis following centrosome loss in RPE1 cells13. Note that USP28∆ has no effect on the mitotic consequences of centrosome loss13 and enables comparison with TRIM37∆ cells, which prevent p53 activation following centrinone treatment by accelerating mitosis in the absence of centrosomes. The gRNA sequence used to target PLK4 exon 5 is depicted, and the efficacy of the inducible knockout in both cell lines was validated by inducing Cas9 expression using doxycycline for four days, followed by sequencing and tracking of indels by decomposition (TIDE) analysis35. Sequence traces show a high frequency of indels, with a 1-bp insertion being the most frequent outcome. h, Protocol used to compare centrinone treatment with the iPLK4 KO in Fig. 2f, g. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 4 Generation of TRIM37 variants, and analysis of the effect of TRIM37 loss on the ubiquitination of histone H2A and transcription.
a, Left, method for generating the cell lines used to test rescue with transgenes encoding wild-type and mutant TRIM37. Centre, top, the point mutations engineered in the ligase and TRAF domains of TRIM37. Right, blot showing the expression of transgene-encoded TRIM37 variants in the pools selected for marker resistance; the percentages of cells expressing the indicated fusions are shown below the blot. b, Structural/sequence analysis used to engineer the TRIM37 TRAF-domain mutant. The USP7 TRAF domain (grey surface) is shown bound to a p53 peptide (cyan stick), with key binding residues W165 and F167 in orange spheres (Protein DataBank (https://www.rcsb.org) code 3MQR). The sequences show the similarity between the peptide-binding pockets of TRIM37 and USP7; the conserved tryptophan (W165 in USP7; W373 in TRIM37) was mutated to alanine to generate the TRIM37 TRAF mutant. c, Images illustrating the effect of expressing wild-type (WT) TRIM37 or engineered variants disrupting ligase activity or TRAF-domain interactions in TRIM37∆ cells. d, Interaction analysis using coexpression of TRIM37 and PLK4 followed by immunoprecipitation of PLK4. WT TRIM37 is expressed at substantially lower levels than ligase-mutant (C18R) TRIM37, suggesting that TRIM37 autoregulates its own stability. The low expression of WT TRIM37 led us to use ligase-mutant TRIM37 for the interaction analysis shown in Fig. 2i. e, Left, immunoblot of H2A conjugated via lysine 119 to ubiquitin (Ub), comparing control and TRIM37∆ RPE1 cells. α-Tubulin (α-tub) served as a loading control. Right, quantification of band intensities indicates that TRIM37 does not reduce ubiquitination of Lys 119 in histone H2A. f, RNA-sequencing (RNA-seq) analysis comparing parental and TRIM37∆ RPE1 cells. A previously defined set of 82 genes encoding centrosomal components43 is marked in red to highlight the lack of change in their mRNA levels. PMID, PubMed identification code. g, RNA-seq analysis comparing parental CHP134 cells with two clones (clones 1 and 2 from Fig. 1f) with substantially lower expression of TRIM37. The centrosome 82-gene set is highlighted in red. h, Lists of genes that are more than twofold downregulated or upregulated. Each of the three test lines (RPE1 TRIM37∆, CHP134 clone 1 and CHP134 clone 2) was compared with the parental line in order to identify genes with statistically significant, more than twofold changes. Cross-comparison of all three test lines and of the two CHP134 clones is summarized in the Venn diagrams; gene names for shared differentially expressed genes are shown below each Venn diagram. For gel source data, see Supplementary Fig. 1. For details on statistics, see Methods; unpaired t-tests assuming equal standard deviation were performed.
Extended Data Fig. 5 Effect of TRIM37 overexpression on the coalescence of pericentriolar material and acentrosomal mitosis.
a, Protocol used to analyse the effect of centrinone treatment on the mitotic dynamics of the CEP192 protein by live imaging (Fig. 3a). b, Immunofluorescence images of centrosome components in DMSO- versus centrinone-treated mitotic RPE1 cells. Scale bar, 10 μm. c, Summary of immunofluorescence analysis, showing which centrosome components were detected in the foci at the poles of acentrosomal spindles in centrinone-treated cells. Scale bar, 10 µm. d, Protocol used to inducibly knock out PLK4 and monitor CEP192 dynamics in mitosis by live imaging (Fig. 3b). e, Protocol used to overexpress TRIM37 and monitor CEP192 dynamics in mitosis by live imaging (Fig. 3c). Note that this is the same cell line used for the analysis in Fig. 3a (no doxycline induction); the analyses in these two conditions were conducted in parallel. f, Additional panels from the time-lapse image sequences shown in Fig. 3b, c for inducible PLK4 knockout and TRIM37 overexpression. Times in minutes after NEBD are noted on each panel. g, RNA-seq analysis comparing two clones that overexpressed TRIM37 with parental RPE1 cells. Elevated TRIM37 transcript levels are evident in both clones. No significant changes in the global transcriptome were otherwise observed. h, Evidence that centrosomes protect CEP192 from TRIM37-dependent degradation. Top, immunoblots of the indicated RPE1 cell lines with and without centrinone treatment. In cells overexpressing TRIM37, centrinone treatment further reduces CEP192 levels; by contrast, in TRIM37∆ cells, centrinone treatment affects CEP192 levels only modestly. Note that RNA-seq analysis indicated no significant change in CEP192 transcript levels between cell lines with varying levels of TRIM37. Bottom, immunoblots of CHP134 parental cells and a derived clone with TRIM37 expression levels roughly 12% of those of the parental cells. Centrinone strongly reduced CEP192 levels in the parental cells but not in the clone with reduced TRIM37 expression. α-Tubulin serves as a loading control. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 6 Evidence that CEP192 is the target of TRIM37 that accounts for enhanced sensitivity to PLK4 inhibition.
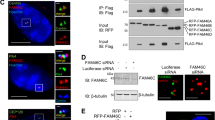
a, Protocol for partial CEP192 inhibition using a short-term inducible knockout, followed by live imaging of mitosis. b, Evidence in CHP134 cells that CEP192 is a functionally important target of TRIM37. A CHP134 clonal cell line with reduced TRIM37 expression (roughly 12% relative to parental CHP134 cells) was stably transduced with a CEP192 shRNA that reduced expression by approximately 75% (immunoblot and quantification below). Right, live imaging of mitosis showed that although reduction of CEP192 levels had no significant effect on the duration of mitosis in DMSO-treated cells, it did significantly extend mitotic duration in centrinone-treated cells. ****P < 0.0001. c, Evidence that the C terminus of CEP192 is ubiquitinated in a TRIM37-dependent manner. The experiment shown in Fig. 4d included co-transfection of HA-tagged ubiquitin. Shown here is the HA–ubiquitin blot (together with FLAG and Myc blots) of the immunoprecipitated C-terminal CEP192 fragment that binds TRIM37. Ubiquitination of this fragment was enhanced in the presence of WT relative to ligase-mutant TRIM37. The FLAG blot shown is the same as in Fig. 4d; the Myc blot is a different exposure of that in Fig. 4d. The other CEP192 fragments are not shown because their stability was affected by coexpression with WT but not ligase-mutant TRIM37, which makes comparisons of ubiquitination profiles difficult. For gel source data, see Supplementary Fig. 1. For details on statistics, see Methods; unpaired t-tests assuming equal standard deviation were performed.
Extended Data Fig. 7 Analysis of CHP134 xenograft tumours on the basis of inducible PLK4 shRNA.
a, Left, generation and characterization of a CHP134 pool with stably integrated inducible PLK4 (iPLK4) shRNA. Following viral transduction, three-day induction of the shRNA with doxycycline followed by immunoblotting and immunofluorescence was used to assess PLK4 depletion and centrosome loss. Right, the immunoblot shows depletion of PLK4 as well as reduction of CEP192 following doxycycline treatment, as is also observed with centrinone; this reduction depended on high TRIM37 expression in CHP134 cells, as it was not observed following induction of PLK4 shRNA in a CHP134-derived line with roughly 10% TRIM37 expression (data not shown). b, Immunofluorescence images showing loss of centrosomes, detected using CEP192, following induction of PLK4 shRNA. Scale bar, 10 μm. c, Quantification of centrosome number after three-day induction of PLK4 shRNA. A longer induction was associated with extensive lethality, as is also observed with centrinone treatment of CHP134 cells. d, Protocol for isolating CHP134 clones with stably integrated iPLK4 shRNA from the pool described in a–c. Right, results of passaging-based analysis, showing that both clones exhibited rapid cessation of proliferation following induction of the shRNA. e, Workflow for tumour xenograft analysis with the two CHP134 iPLK4-shRNA clones. Tumours were generated in female BALB/c nude mice, and the shRNA was induced by switching to a doxycycline-containing diet. Tumour volume and body weight were measured over time after induction. f, Time course of tumour growth in BALB/c nude mice for the two CHP134 PLK4 shRNA clonal lines following induction of shRNA (doxycycline) versus no induction (control). Error bars show standard error of the mean. Statistical significance was evaluated using unpaired t-tests (Welch’s correction). For gel source data, see Supplementary Fig. 1.
Supplementary information
Supplementary Figure 1
Uncropped images of blots presented in the main paper and Extended Data.
Supplementary Video 1
Mitosis in control DMSO-treated RPE1 and centrinone-treated RPE1, TRIM37∆ and TRIM37-overexpressing (TetOn-TRIM37 clone 5) cells. Cells were treated for 2 d with DMSO or centrinone and doxycycline was added for 24h before acquisition of a 5 × 2-µm confocal z-series every 4 min for 12 h. DNA was labeled with SiR-DNA. A representative example is shown for each condition. Images are maximum-intensity projections. Images were acquired on a CQ1 spinning disk confocal system (Yokogawa Electric Corporation) equipped with a 40× (0.95 NA) U-PlanApo objective and 2,560 × 2,160-pixel sCMOS camera (Andor Technology). Playback rate is 1,200× real time (5 frames/s).
Supplementary Video 2
Mitosis in DMSO- and centrinone- treated RPE1 and CHP134 cells. Cells were treated for three cell cycle durations with DMSO or centrinone before acquisition of a 5 × 2-µm confocal z-series every 4 min for 12 h. Cells are labeled with H2b-RFP. Two representative examples are shown for each condition. Images are maximum-intensity projections. Images were acquired on a CQ1 spinning disk confocal system (Yokogawa Electric Corporation) equipped with a 40× (0.95 NA) U-PlanApo objective and 2,560 × 2,160-pixel sCMOS camera (Andor Technology). Playback rate is 1,200× real time (5 frames/s).
Source data
Rights and permissions
About this article
Cite this article
Meitinger, F., Ohta, M., Lee, KY. et al. TRIM37 controls cancer-specific vulnerability to PLK4 inhibition. Nature 585, 440–446 (2020). https://doi.org/10.1038/s41586-020-2710-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2710-1
This article is cited by
-
RIT1 regulates mitosis and promotes proliferation by interacting with SMC3 and PDS5 in hepatocellular carcinoma
Journal of Experimental & Clinical Cancer Research (2023)
-
TRIM37 exacerbates hepatic ischemia/reperfusion injury by facilitating IKKγ translocation
Molecular Medicine (2023)
-
Spindle function and Wnt pathway inhibition by PBX1 to suppress tumor progression via downregulating DCDC2 in colorectal cancer
Oncogenesis (2023)
-
The TRIM37 variants in Mulibrey nanism patients paralyze follicular helper T cell differentiation
Cell Discovery (2023)
-
An updated view on the centrosome as a cell cycle regulator
Cell Division (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.