Abstract
Leishmaniasis refers to a complex of diseases, caused by the intracellular parasitic protozoans belonging to the genus Leishmania. Among the three types of disease manifestations, the most severe type is visceral leishmaniasis, which is caused by Leishmania donovani, and is diagnosed in more than 20,000 cases annually, worldwide. Because the current therapeutic options for disease treatment are associated with several limitations, the identification of new potential leads/drugs remains necessary. In this study, a combined approach was used, based on two different virtual screening (VS) methods, which were designed to select promising antileishmanial agents from among the entire sesquiterpene lactone (SL) dataset registered in SistematX, a web interface for managing a secondary metabolite database that is accessible by multiple platforms on the Internet. Thus, a ChEMBL dataset, including 3159 and 1569 structures that were previously tested against L. donovani amastigotes and promastigotes in vitro, respectively, was used to develop two random forest models, which performed with greater than 74% accuracy in both the cross-validation and test sets. Subsequently, a ligand-based VS assay was performed against the 1306 SistematX-registered SLs. In parallel, the crystal structures of three L. donovani target proteins, N-myristoyltransferase, ornithine decarboxylase, and mitogen-activated protein kinase 3, and a homology model of pteridine reductase 1 were used to perform a structure-based VS, using molecular docking, of the entire SistematX SL dataset. The consensus analysis of these two VS approaches resulted in the normalization of probability scores and identified 13 promising, enzyme-targeting, antileishmanial SLs from SistematX that may act against L. donovani.
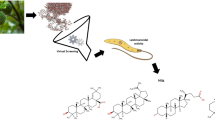
Graphic abstract
A combined approach based on two different virtual screening methods (structure-based and ligand-based) was performed using an in-house dataset composed of 1306 sesquiterpene lactones to identify potential antileishmanial (Leishmania donovani) structures.









Similar content being viewed by others
References
Babuadze G, Farlow J, de Koning HP, Carrillo E, Chakhunashvili G, Murskvaladze M, Kekelidze M, Karseladze I, Kokaia N, Kalandadze I, Tsereteli D, Markhvashvili I, Sidamonidze K, Chanturia G, Adeishvili E, Imnadze P (2016) Seroepidemiology and molecular diversity of Leishmania donovani complex in Georgia. Parasit Vectors 9(1):279. https://doi.org/10.1186/s13071-016-1558-6
Wijerathna T, Gunathunga S, Gunathilaka N (2020) Recent developments and future directions in the paratransgenesis based control of Leishmania transmission. Biol Control. https://doi.org/10.1016/j.biocontrol.2020.104260
Basano SdA, Camargo LMA (2004) Leishmaniose tegumentar americana: histórico, epidemiologia e perspectivas de controle. Rev Bras Epidemiol. https://doi.org/10.1590/S1415-790X2004000300010
Fiocruz As Leishmanioses. https://www.dbbm.fiocruz.br/tropical/leishman/leishext/html/ciclo_biol_gico.htm. Accessed 29 Feb 2020
Ponte-Sucre A, Gamarro F, Dujardin JC, Barrett MP, Lopez-Velez R, Garcia-Hernandez R, Pountain AW, Mwenechanya R, Papadopoulou B (2017) Drug resistance and treatment failure in leishmaniasis: a 21st century challenge. PLoS Negl Trop Dis 11(12):e0006052. https://doi.org/10.1371/journal.pntd.0006052
Kumar A (2020) Survival strategies of Leishmania parasite: too many questions and few answers. Curr Pharmacol Rep. https://doi.org/10.1007/s40495-020-00209-6
Bhattacharya P, Dey R, Dagur PK, Joshi AB, Ismail N, Gannavaram S, Debrabant A, Akue AD, KuKuruga MA, Selvapandiyan A, McCoy JP Jr, Nakhasi HL (2016) Live attenuated Leishmania donovani centrin knock out parasites generate non-inferior protective immune response in aged mice against visceral Leishmaniasis. PLoS Negl Trop Dis 10(8):e0004963. https://doi.org/10.1371/journal.pntd.0004963
WHO World Health Organization. Leishmaniasis: epidemiological situation. https://www.who.int/leishmaniasis/burden/en/. Accessed 09 Mar 2020
Anversa L, Tiburcio MGS, Richini-Pereira VB, Ramirez LE (2018) Human leishmaniasis in Brazil: a general review. Rev Assoc Med Bras (1992) 64(3):281–289. https://doi.org/10.1590/1806-9282.64.03.281
Alvar J, Velez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M, Team WHOLC (2012) Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE 7(5):e35671. https://doi.org/10.1371/journal.pone.0035671
Hoet S, Opperdoes F, Brun R, Quetin-Leclercq J (2004) Natural products active against African trypanosomes: a step towards new drugs. Nat Prod Rep 21(3):353–364. https://doi.org/10.1039/b311021b
Utzinger J, Becker SL, Knopp S, Blum J, Neumayr AL, Keiser J, Hatz CF (2012) Neglected tropical diseases: diagnosis, clinical management, treatment and control. Swiss Med Wkly 142:w13727. https://doi.org/10.4414/smw.2012.13727
Gilbert IH (2013) Drug discovery for neglected diseases: molecular target-based and phenotypic approaches: miniperspectives series on phenotypic screening for antiinfective targets. J Med Chem 56(20):7719–7726. https://doi.org/10.1021/jm400362b
Palma LC, Ferreira L, Petersen A, Dias BRS, Menezes JPB, Moreira DRM, Hernandes MZ, Veras PST (2019) A docking-based structural analysis of geldanamycin-derived inhibitor binding to human or Leishmania Hsp90. Sci Rep 9(1):14756. https://doi.org/10.1038/s41598-019-51239-0
Newman DJ, Cragg GM (2020) Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod 83(3):770–803. https://doi.org/10.1021/acs.jnatprod.9b01285
Herrera Acevedo C, Scotti L, Feitosa Alves M, Formiga Melo Diniz M, Scotti M (2017) Computer-aided drug design using sesquiterpene lactones as sources of new structures with potential activity against infectious neglected diseases. Molecules 22(1):79. https://doi.org/10.3390/molecules22010079
Schmidt TJ, Nour AM, Khalid SA, Kaiser M, Brun R (2009) Quantitative structure-antiprotozoal activity relationships of sesquiterpene lactones. Molecules 14(6):2062–2076. https://doi.org/10.3390/molecules14062062
Karioti A, Skaltsa H, Kaiser M, Tasdemir D (2009) Trypanocidal, leishmanicidal and cytotoxic effects of anthecotulide-type linear sesquiterpene lactones from Anthemis auriculata. Phytomedicine 16(8):783–787. https://doi.org/10.1016/j.phymed.2008.12.008
Kumar P, Kumar A, Verma SS, Dwivedi N, Singh N, Siddiqi MI, Tripathi RP, Dube A, Singh N (2008) Leishmania donovani pteridine reductase 1: biochemical properties and structure-modeling studies. Exp Parasitol 120(1):73–79. https://doi.org/10.1016/j.exppara.2008.05.005
Saudagar P, Saha P, Saikia AK, Dubey VK (2013) Molecular mechanism underlying antileishmanial effect of oxabicyclo[3.3.1]nonanones: inhibition of key redox enzymes of the pathogen. Eur J Pharm Biopharm 85(3 Pt A):569–577. https://doi.org/10.1016/j.ejpb.2013.08.014
Shukla AK, Patra S, Dubey VK (2012) Iridoid glucosides from Nyctanthes arbortristis result in increased reactive oxygen species and cellular redox homeostasis imbalance in Leishmania parasite. Eur J Med Chem 54:49–58. https://doi.org/10.1016/j.ejmech.2012.04.034
Shukla AK, Patra S, Dubey VK (2011) Evaluation of selected antitumor agents as subversive substrate and potential inhibitor of trypanothione reductase: an alternative approach for chemotherapy of Leishmaniasis. Mol Cell Biochem 352(1–2):261–270. https://doi.org/10.1007/s11010-011-0762-0
Saudagar P, Dubey VK (2011) Cloning, expression, characterization and inhibition studies on trypanothione synthetase, a drug target enzyme, from Leishmania donovani. Biol Chem 392(12):1113–1122. https://doi.org/10.1515/BC.2011.222
Resh MD (1999) Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta (BBA) Mol Cell Res 1451(1):1–16. https://doi.org/10.1016/S0167-4889(99)00075-0
Tate EW, Bell AS, Rackham MD, Wright MH (2014) N-Myristoyltransferase as a potential drug target in malaria and leishmaniasis. Parasitology 141(1):37–49. https://doi.org/10.1017/S0031182013000450
Chawla B, Madhubala R (2010) Drug targets in Leishmania. J Parasit Dis 34(1):1–13. https://doi.org/10.1007/s12639-010-0006-3
Nichol CA, Smith GK, Duch DS (1985) Biosynthesis and metabolism of tetrahydrobiopterin and molybdopterin. Annu Rev Biochem 54(1):729–764. https://doi.org/10.1146/annurev.bi.54.070185.003501
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Noble WS (2006) What is a support vector machine? Nat Biotechnol 24(12):1565–1567. https://doi.org/10.1038/nbt1206-1565
Fourches D, Muratov E, Tropsha A (2015) Curation of chemogenomics data. Nat Chem Biol 11(8):535. https://doi.org/10.1038/nchembio.1881
Muratov EN, Varlamova EV, Artemenko AG, Polishchuk PG, Kuz'min VE (2012) Existing and developing approaches for QSAR analysis of mixtures. Mol Inform 31(3–4):202–221. https://doi.org/10.1002/minf.201100129
Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, Dearden J, Gramatica P, Martin YC, Todeschini R (2014) QSAR modeling: where have you been? Where are you going to? J Med Chem 57(12):4977–5010. https://doi.org/10.1021/jm4004285
Cruciani G, Crivori P, Carrupt P-A, Testa B (2000) Molecular fields in quantitative structure–permeation relationships: the VolSurf approach. J Mol Struct (Thoechem) 503(1):17–30. https://doi.org/10.1016/S0166-1280(99)00360-7
Cruciani G, Pastor M, Guba W (2000) VolSurf: a new tool for the pharmacokinetic optimization of lead compounds. Eur J Pharm Sci 11(Suppl 2):S29–39. https://doi.org/10.1016/s0928-0987(00)00162-7
Berthold MR, Cebron N, Dill F, Gabriel TR, Kötter T, Meinl T, Ohl P, Thiel K, Wiswedel B (2009) KNIME-the Konstanz information miner: version 2.0 and beyond. ACM SIGKDD Explor Newslett 11(1):26–31. https://doi.org/10.1145/1656274.1656280
Fourches D, Pu D, Tassa C, Weissleder R, Shaw SY, Mumper RJ, Tropsha A (2010) Quantitative nanostructure−activity relationship modeling. ACS Nano 4(10):5703–5712
Hanley JA, McNeil BJ (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143(1):29–36. https://doi.org/10.1148/radiology.143.1.7063747
Matthews BW (1975) Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta 405(2):442–451. https://doi.org/10.1016/0005-2795(75)90109-9
Baell JB, Holloway GA (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 53(7):2719–2740. https://doi.org/10.1021/jm901137j
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2012) GenBank. Nucleic Acids Res 41(D1):D36–D42. https://doi.org/10.1093/nar/gkq1079
Bernstein FC, Koetzle TF, Williams GJ, Meyer EF Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M (1978) The Protein Data Bank: a computer-based archival file for macromolecular structures. Arch Biochem Biophys 185(2):584–591. https://doi.org/10.1016/0003-9861(78)90204-7
Gourley DG, Schuttelkopf AW, Leonard GA, Luba J, Hardy LW, Beverley SM, Hunter WN (2001) Pteridine reductase mechanism correlates pterin metabolism with drug resistance in trypanosomatid parasites. Nat Struct Biol 8(6):521–525. https://doi.org/10.1038/88584
Šali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234(3):779–815. https://doi.org/10.1006/jmbi.1993.1626
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26(2):283–291. https://doi.org/10.1107/S0021889892009944
Lovell SC, Davis IW, Arendall WB, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC (2003) Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins Struct Funct Bioinform 50(3):437–450. https://doi.org/10.1002/prot.10286
Diego S (2007) Discovery studio modeling environment release 2.1. Accelerys Software Inc, San Diego
Dufe VT, Ingner D, Heby O, Khomutov AR, Persson L, Al-Karadaghi S (2007) A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane. Biochem J 405(2):261–268. https://doi.org/10.1042/BJ20070188
Brannigan JA, Smith BA, Yu Z, Brzozowski AM, Hodgkinson MR, Maroof A, Price HP, Meier F, Leatherbarrow RJ, Tate EW, Smith DF, Wilkinson AJ (2010) N-myristoyltransferase from Leishmania donovani: structural and functional characterisation of a potential drug target for visceral leishmaniasis. J Mol Biol 396(4):985–999. https://doi.org/10.1016/j.jmb.2009.12.032
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Berendsen HJ, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91(1–3):43–56. https://doi.org/10.1016/0010-4655(95)00042-E
Schuttelkopf AWVAD (2004) PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr D Biol Crystallogr. https://doi.org/10.1107/S0907444904011679
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612. https://doi.org/10.1002/jcc.20084
Kumari R, Kumar R, Open Source Drug Discovery C, Lynn A (2014) g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962
Castillo-González D, Mergny J-L, De Rache A, Pérez-Machado G, Cabrera-Pérez MA, Nicolotti O, Introcaso A, Mangiatordi GF, Guédin A, Bourdoncle A (2015) Harmonization of QSAR best practices and molecular docking provides an efficient virtual screening tool for discovering new G-quadruplex ligands. J Chem Inf Model 55(10):2094–2110. https://doi.org/10.1021/acs.jcim.5b00415
Rueda M, Abagyan R (2016) Best practices in docking and activity prediction. bioRxiv. https://doi.org/10.1101/039446
Scotti MT, Herrera-Acevedo C, Oliveira TB, Costa RPO, Santos S, Rodrigues RP, Scotti L, Da-Costa FB (2018) SistematX, an online web-based cheminformatics tool for data management of secondary metabolites. Molecules 23(1):103. https://doi.org/10.3390/molecules23010103
Geissman T, Griffin S, Waddell TG, Chen HH (1969) Sesquiterpene lactones. some new constituents of Ambrosia species: A. psilostachya and A. acanthicarpa. Phytochemistry 8(1):145–150. https://doi.org/10.1016/S0031-9422(00)85806-9
Schmidt TJ (1999) Toxic activities of sesquiterpene lactones: structural and biochemical aspects. Curr Org Chem 3(577–608):4
Popławski J, Holub M, Samek Z, Herout V (1971) On terpenes. CCIX. Arnicolides-sesquiterpenic lactones from the leaves of Arnica montana L. Collect Czechoslov Chem Commun 36(6):2189–2199. https://doi.org/10.1135/cccc19712189
Xie C, Wang H, Sun X, Meng L, Wang M, Bartlam M, Guo Y (2015) Isolation, characterization, and antiproliferative activities of eudesmanolide derivatives from the flowers of Inula japonica. J Agric Food Chem 63(41):9006–9011. https://doi.org/10.1021/acs.jafc.5b03075
Acevedo CH, Scotti L, Scotti MT (2018) In silico studies designed to select sesquiterpene lactones with potential antichagasic activity from an in-house Asteraceae database. ChemMedChem 13(6):634–645. https://doi.org/10.1002/cmdc.201700743
Walker RH, Wells LW, McGUIRE JA (1989) Bristly starbur (Acanthospermum hispidum) interference in peanuts (Arachis hypogaea). Weed Sci 37(2):196–200. https://doi.org/10.1017/S0043174500071770
Kaur J, Dube D, Ramachandran R, Singh P, Singh N (2012) Thianthrene is a novel inhibitor of Leishmania donovani pteridine reductase 1 (PTR1). J Mol Biochem 1(2):68–75
Rozo-Lugo C, Cuca-Suárez LE, Schmidt TJ, Coy-Barrera E (2018) Tetrahydrobenzofuran-6 (2 H)-one Neolignans from Ocotea heterochroma: their platelet activating factor (PAF) antagonistic activity and in silico insights into the PAF receptor binding mode. J Nat Prod 81(9):1968–1975. https://doi.org/10.1021/acs.jnatprod.8b00189
Acknowledgements
We thank the CNPq and Capes for financial Support, Grant Numbers 309648/2019-0 and 431254/2018-4, and the Student Agreement Program of Graduate—PEC-PG of CNPq—Brazil. UMNG is also thanked for the financial support through Project IMP-CIAS-2924. This study was performed as part of the activities of the Research Network Natural Products against Neglected Diseases (ResNet-NPND), https://resnetnpnd.org/.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Herrera-Acevedo, C., Dos Santos Maia, M., Cavalcanti, É.B.V.S. et al. Selection of antileishmanial sesquiterpene lactones from SistematX database using a combined ligand-/structure-based virtual screening approach. Mol Divers 25, 2411–2427 (2021). https://doi.org/10.1007/s11030-020-10139-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-020-10139-6