Abstract
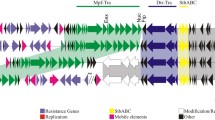
Genome sequence of Acinetobacter baumannii DS002 revealed the existence of seven contigs with features of indigenous plasmids. Of the seven contigs, three of them have shown size and sequence identity. They appeared to have been generated due to the unique recombination events leading to a large-scale recombination and sequence inversions. The rest of the indigenous plasmids have shown significant size variations and contained the genetic repertoire required for the detoxification of formaldehyde and biosynthesis of exopolysaccharides. Genetic modules encoding novel toxin–antitoxin systems were found in most of the plasmids to ensure their survival in the host. In some instances, the toxin and antitoxin coding sequences were found on two different plasmids promoting the cosegregation of these two plasmids into the daughter cells.






Similar content being viewed by others
References
Alikhan N. F., Petty N. K., Ben Zakour N. L. and Beatson S. A. 2011 BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402.
Alvarez-Martinez C. E. and Christie P. J. 2009 Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73, 775–808.
Aminov R. I. 2011 Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2, 158.
Antunes L. C., Visca P. and Towner K. J. 2014 Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 71, 292–301.
Balaji V., Rajenderan S., Anandan S. and Biswas I. 2015 Genome sequences of two multidrug-resistant acinetobacter baumannii clinical strains isolated from Southern India. Genome Announc. 3. e01010–e01015.
Bates S., Cashmore A. M. and Wilkins B. M. 1998 IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae: involvement of the tra2 mating system. J. Bacteriol. 180, 6538–6543.
Bochtler M., Kolano A. and Xu G. L. 2017 DNA demethylation pathways: additional players and regulators. BioEssays 39, 1–13.
Carattoli A. 2009 Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238.
Chen K., Reuter M., Sanghvi B., Roberts G. A., Cooper L. P., Tilling M. et al. 2014 ArdA proteins from different mobile genetic elements can bind to the EcoKI Type I DNA methyltransferase of E. coli K12. Biochi. Biophy. Acta 1844, 505–511.
Chen N. H., Djoko K. Y., Veyrier F. J., and McEwan A. G. 2016 Formaldehyde stress responses in bacterial pathogens. Front. Microbiol. 7, 257.
Clemmer K. M., Bonomo R. A. and Rather P. N. 2011 Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157, 2534–2544.
Dai Z., Dong H., Zhang Y. and Li Y. 2016 Elucidating the contributions of multiple aldehyde/alcohol dehydrogenases to butanol and ethanol production in Clostridium acetobutylicum. Sci. Rep. 6, 28189.
Darmon E. and Leach D. R. 2014 Bacterial genome instability. Microbiol. Mol. Biol. Rev. 78, 1–39.
del Solar G., Giraldo R., Ruiz-Echevarria M. J., Espinosa M. and Diaz-Orejas R. 1998 Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62, 434–464.
Dimitriu T., Misevic D., Lotton C., Brown S. P., Lindner A. B. and Taddei F. 2016 Indirect fitness benefits enable the spread of host genes promoting costly transfer of beneficial plasmids. PLoS Biol. 14, e1002478.
Disque-Kochem C. and Dreiseikelmann B. 1997 The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J. Bacteriol. 179, 6133–6137.
Fukushima T., Yao Y., Kitajima T., Yamamoto H. and Sekiguchi J. 2007 Characterization of new L,D-endopeptidase gene product CwlK (previous YcdD) that hydrolyzes peptidoglycan in Bacillus subtilis. Mol. Genet. Genom. 278, 371–383.
Funnell B. and Phillips G. J. (ed.). 2004 In Plasmid biology. ASM Press, Washington.
Gonzalez C. F., Proudfoot M., Brown G., Korniyenko Y., Mori H., Savchenko A. V. et al. 2006 Molecular basis of formaldehyde detoxification. Characterization of two S-formylglutathione hydrolases from Escherichia coli, FrmB and YeiG. J. Biol. Chem. 281, 14514–14522.
Gray M. J., Wholey W. Y., Parker B. W., Kim M. and Jakob U. 2013 NemR is a bleach-sensing transcription factor. J. Biol. Chem. 288, 13789–13798.
Groisman E. A. and Ochman H. 1996 Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87, 791–794.
Heinemann J. A. 1991 Genetics of gene transfer between species. Trends Genet. 7, 181–185.
Hyatt D., Chen G. L., Locascio P. F., Land M. L., Larimer F. W. and Hauser L. J. 2010 Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformat. 11, 119.
Johnson T. J. and Nolan L. K. 2009 Plasmid replicon typing. Methods Mol. Biol. 551, 27–35.
Khiste N. and Ilie L. 2017 HISEA: HIerarchical SEed Aligner for PacBio data. BMC Bioinformat. 18, 564.
Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H. and Phillippy A. M. 2017 Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736.
Kwong S. M., Ramsay J. P., Jensen S. O. and Firth N. 2017 Replication of Staphylococcal resistance plasmids. Front. Microbiol. 8, 2279.
Lamers R. P., Nguyen U. T., Nguyen Y., Buensuceso R. N. and Burrows L. L. 2015 Loss of membrane-bound lytic transglycosylases increases outer membrane permeability and beta-lactam sensitivity in Pseudomonas aeruginosa. Microbiologyopen 4, 879–895.
Lee P. A., Tullman-Ercek D. and Georgiou G. 2006 The bacterial twin-arginine translocation pathway. Annu. Rev. Microbiol. 60, 373–395.
Lipps G. E. 2008 Plasmids: current research and future trends. Horizon Scientific Press.
Longkumer T., Kamireddy S., Muthyala V. R., Akbarpasha S., Pitchika, G. K., Kodetham G. et al. 2013 Acinetobacter phage genome is similar to Sphinx 2.36, the circular DNA copurified with TSE infected particles. Sci. Rep. 3, 2240.
MacLean R. C. and San Millan, A. 2015 Microbial evolution: towards resolving the plasmid paradox. Curr. Biol. 25, R764–R767.
Marx C. J., Chistoserdova L. and Lidstrom M. E. 2003 Formaldehyde-detoxifying role of the tetrahydromethanopterin-linked pathway in Methylobacterium extorquens AM1. J. Bacteriol. 185, 7160–7168.
Mesnage S. and Fouet A. 2002. Plasmid-encoded autolysin in Bacillus anthracis: modular structure and catalytic properties. J. Bacteriol. 184, 331–334.
Nekrasov S. V., Agafonova O. V., Belogurova N. G., Delver E. P. and Belogurov A. A. 2007 Plasmid-encoded antirestriction protein ArdA can discriminate between type I methyltransferase and complete restriction-modification system. J. Mol. Biol. 365, 284–297.
Ohtsubo Y., Ikeda-Ohtsubo W., Nagata Y. and Tsuda M. 2008 Genome matcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics 9, 376.
Orlek A., Stoesser N., Anjum M. F., Doumith M., Ellington M. J., Peto T. et al. 2017 Plasmid classification in an era of whole-genome sequencing: application in studies of antibiotic resistance epidemiology. Front. Microbiol. 8, 182.
Pallen M. J. and Wren B. W. 1997 The HtrA family of serine proteases. Mol. Microbiol. 26, 209–221.
Peleg A. Y., Seifert H. and Paterson D. L. 2008 Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582.
Parales R. E. and Resnick S. M. 2004 Aromatic hydrocarbon dioxygenases. In Biodegradation and bioremediation. soil biology (ed. A. Singh and O. P. Ward), vol 2. Springer, Berlin, Heidelberg.
Rose R. W., Bruser T., Kissinger J. C. and Pohlschroder M. 2002 Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45, 943–950.
Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069.
Shintani M., Sanchez Z. K. and Kimbara K. 2015 Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol. 6, 242.
Smillie C., Garcillan-Barcia M. P., Francia M. V., Rocha E. P. and de la Cruz F. 2010 Mobility of plasmids. Microbiol. Molecular Biol. Rev. 74, 434–452.
Spero M. A., Aylward F. O., Currie C. R. and Donohue T. J. 2015 Phylogenomic analysis and predicted physiological role of the proton-translocating NADH:quinone oxidoreductase (complex I) across bacteria. mBio 6. e00389-15.
Stingele F., Newell J. W. and Neeser J. R. 1999 Unraveling the function of glycosyltransferases in Streptococcus thermophilus Sfi6. J. Bacteriol. 181, 6354–6360.
Suzuki H., Yano H., Brown C. J. and Top E. M. 2010 Predicting plasmid promiscuity based on genomic signature. J. Bacteriol. 192, 6045–6055.
Vijaykumar S., Balaji V. and Biswas I. 2015 Complete genome sequence of Acinetobacter baumannii strain B8300, which displays high twitching motility. Genome Announc. 3.
Vollmer W., Joris B., Charlier P. and Foster S. 2008 Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286.
Wong J. J., Lu J. and Glover J. N. 2012 Relaxosome function and conjugation regulation in F-like plasmids - a structural biology perspective. Mol. Microbiol. 85, 602–617.
Yakkala H., Samantarrai D., Gribskov M. and Siddavattam D. 2019 Comparative genome analysis reveals niche-specific genome expansion in Acinetobacter baumannii strains. PLoS One 14, e0218204.
Acknowledgments
DS received Research Associateship from a CSIR, New Delhi sponsored research Project (37(1684)17/EMT-11). YS is a recipient of Research fellowship from a DST sponsored women scientist grant (SR/WOS-A/LS-1225/2015). Science, Engineering Research Board (SERB), Department of Science Technology, New Delhi supported the research work in DS laboratory.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: H. A. Ranganath
Rights and permissions
About this article
Cite this article
Samantarrai, D., Yakkala, H. & Siddavattam, D. Analysis of indigenous plasmid sequences of A. baumannii DS002 reveals the existence of lateral mobility and extensive genetic recombination among Acinetobacter plasmids. J Genet 99, 71 (2020). https://doi.org/10.1007/s12041-020-01232-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-020-01232-8