Gene Expression Profile at the Motor Endplate of the Neuromuscular Junction of Fast-Twitch Muscle
- 1Division of Neurogenetics, Center for Neurological Diseases and Cancer, Nagoya University Graduate School of Medicine, Nagoya, Japan
- 2Department of Genetics, Research Institute of Environmental Medicine (RIeM), Nagoya University, Nagoya, Japan
The neuromuscular junction (NMJ) is a prototypic chemical synapse between the spinal motor neuron and the motor endplate. Gene expression profiles of the motor endplate are not fully elucidated. Collagen Q (ColQ) is a collagenic tail subunit of asymmetric forms of acetylcholinesterase and is driven by two distinct promoters. pColQ1 is active throughout the slow-twitch muscle, whereas pColQ1a is active at the motor endplate of fast-twitch muscle. We made a transgenic mouse line that expresses nuclear localization signal (NLS)-attached Cre recombinase under the control of pColQ1a (pColQ1a-Cre mouse). RiboTag mouse expresses an HA-tagged ribosomal subunit, RPL22, in cells expressing Cre recombinase. We generated pColQ1a-Cre:RiboTag mouse, and confirmed that HA-tagged RPL22 was enriched at the NMJ of tibialis anterior (TA) muscle. Next, we confirmed that Chrne and Musk that are specifically expressed at the NMJ were indeed enriched in HA-immunoprecipitated (IP) RNA, whereas Sox10 and S100b, markers for Schwann cells, and Icam1, a marker for vascular endothelial cells, and Pax3, a marker for muscle satellite cells, were scarcely detected. Gene set enrichment analysis (GSEA) of RNA-seq data showed that “phosphatidylinositol signaling system” and “extracellular matrix receptor interaction” were enriched at the motor endplate. Subsequent analysis revealed that genes encoding diacylglycerol kinases, phosphatidylinositol kinases, phospholipases, integrins, and laminins were enriched at the motor endplate. We first characterized the gene expression profile under translation at the motor endplate of TA muscle using the RiboTag technique. We expect that our gene expression profiling will help elucidate molecular mechanisms of the development, maintenance, and pathology of the NMJ.
Introduction
Neuromuscular junction (NMJ) is a prototypic synapse between the spinal motor neuron and the motor endplate (Hirsch, 2007; Ohno et al., 2017). At the NMJ, acetylcholinesterase (AChE) rapidly hydrolyzes acetylcholine (ACh) released from the nerve terminal (Ohno et al., 1998; Gaspersic et al., 1999). At the NMJ, a globular form of AChE makes a homomeric tetramer. One, two, and three tetrameric AChE are bound to triple-helical collagen Q (ColQ) to make asymmetric A4, A8, and A12 forms, and are anchored to the synaptic basal lamina (Nazim et al., 2017). Transcription of Colq is driven by two distinct promoters of pColQ1 and pColQ1a (Figure 1A; Krejci et al., 1999; Lee et al., 2004; Ting et al., 2005). pColQ1 is active throughout the slow-twitch muscle. pColQ1 starts transcription from Colq exon 1 to generate A4 and A8 forms. In contrast, pColQ1a is active at the NMJ, but not at the extrajunctional regions, of the fast-twitch muscle. pColQ1a starts transcription from Colq exon 1a to predominantly generate A12 form. Human pColQ1a carries consensus sequences for transcriptional factors E-protein (E-box, CANNTG), NFAT (GGAAA), c-Ets transcription factor [c-Ets, (C/A)GGA(A/T)], Elk-1, N-box (CCGGAA), and MEF2 (CTAAAAATAA), which play essential roles in muscle-specific and NMJ-specific transcriptional activities (Lee et al., 2004).
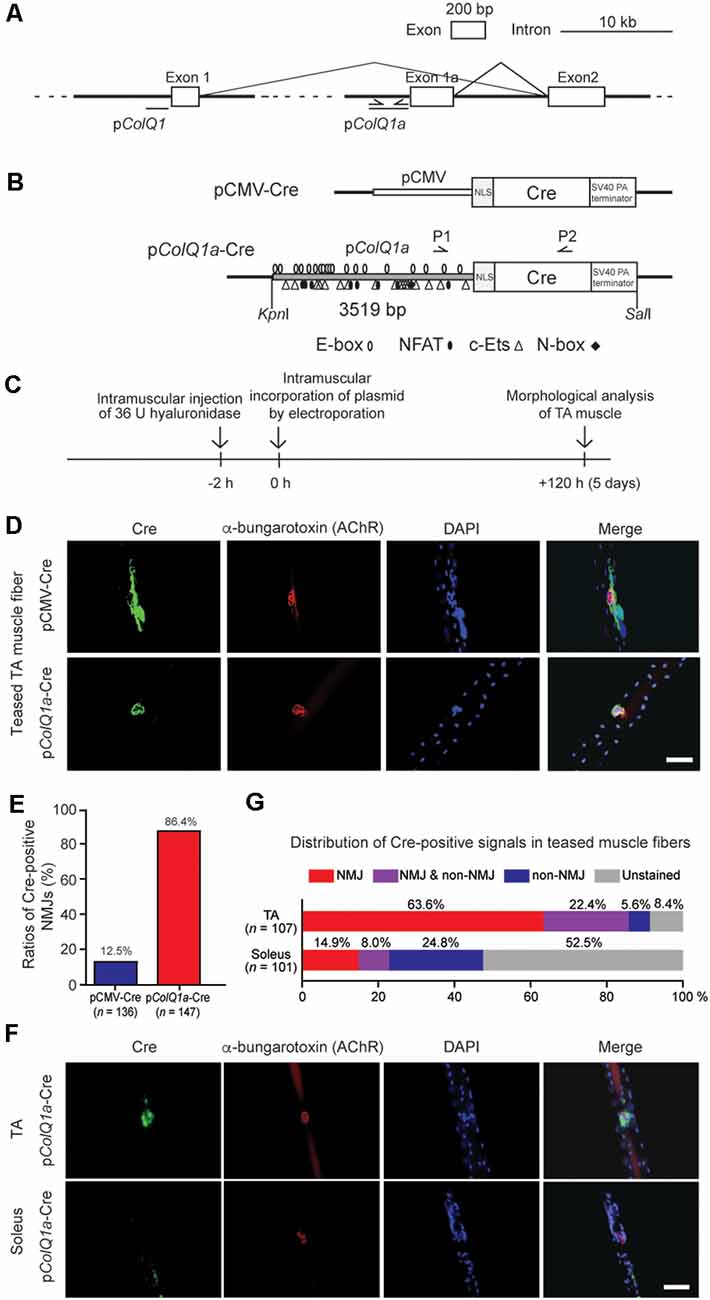
Figure 1. Mouse pColQ1a promoter expresses Cre at the neuromuscular junction (NMJ) of the tibialis anterior (TA) muscle. (A) Mouse Colq is driven by two distinct promoters. pColQ1 drives transcription from exon 1, whereas pColQ1a drives transcription from exon 1a. PCR primers (Supplementary Table S1) to amplify a 3519-bp fragment of pColQ1a are indicated by arrows. (B) Schematic of pCMV-Cre and pColQ1a-Cre. pCMV or pColQ1a was fused to the nuclear localization signal (NLS) sequence and the Cre recombinase gene. P1 and P2 arrows indicate the positions of genotyping primers. Putative binding motifs of transcription factors (E-box, NFAT, c-Ets, and N-box) are indicated (Lee et al., 2004). (C) The protocol of muscle electroporation. (D) Representative immunostaining of Cre (green) and Alexa594-conjugated α-bungarotoxin-staining for AChR (red) of teased TA muscle fibers at 5 days after electroporation of the indicated plasmids. The nuclei were stained with DAPI. Scale bar = 30 μm. (E) Ratios of Cre-positive NMJs in teased TA muscle fibers electroporated with pCMV-Cre and pColQ1a-Cre in 4 pCMV-Cre mice (n = 136 fibers, respectively) and 5 pColQ1a-Cre mice (n = 147 fibers). P < 0.001 by Fisher’s exact test (see Supplementary Table S3 for individual counts). (F) Representative images showing NMJ-specific expression of Cre in teased TA muscle fibers but not in teased soleus muscle fibers of the pColQ1a-Cre mouse. Teased muscle fibers were stained with anti-Cre antibody (green) and Alexa594-conjugated α-bungarotoxin 594 (red). Nuclei were stained with DAPI. Scale bar = 30 μm. (G) Distribution of Cre-positive signals along teased muscle fibers of TA and soleus muscles of the pColQ1a-Cre mouse. The total number of teased muscle fibers is shown in parentheses. P < 0.001 by Fisher’s exact test (see Supplementary Table S4 for individual counts).
Massive parallel sequencing of RNA (RNA-seq) enabled the identification of gene expression profiles in specific tissues and cells at an unprecedented level of accuracy. Also, tissue-specific and cell-type-specific transcriptomic profiling is made available by fluorescence-activated cell sorting (FACS), laser capture microdissection, and single-cell RNA-seq (scRNA-seq; Kim et al., 2015). However, gene expression profiles can be artificially changed in the course of FACS and scRNA-seq procedures. In laser capture microdissection, target cells are frequently contaminated by surrounding cells, because target cells should be free from laser burns. RiboTag mouse technique was developed to circumvent these disadvantages (Sanz et al., 2009). RiboTag mouse carries an RPL22 subunit of the ribosome with a hemagglutinin tag at the C-terminal end (RPL22-HA), and its expression is induced by Cre recombinase (Sanz et al., 2009). Cell-type-specific expression of Cre recombinase thus enables isolation of total mRNA under translation in a cell-type-specific manner.
In our study, we first generated a transgenic pColQ1a-Cre mouse that specifically expresses Cre recombinase in the subsynaptic nuclei of fast-twitch muscle. As far as we know, no transgenic mouse has been generated to express Cre recombinase at the motor endplate. We confirmed that Cre recombinase is highly expressed at the NMJ of fast-twitch muscle, but not of slow-twitch muscle, in the pColQ1a-Cre mouse. To characterize gene expression profiles at the motor endplate of fast-twitch muscle, we made pColQ1a-Cre:RiboTag mouse that expresses RPL22-HA only at the motor endplate. Quantitative RT-PCR (qRT-PCR) showed that mRNA immunoprecipitated with the HA tag from tibialis anterior (TA) muscle of pColQ1a-Cre:RiboTag mouse was enriched for motor endplate-specific genes, and mostly depleted for genes specific for Schwann cells and vascular endothelial cells. RNA-seq analysis of HA-immunoprecipitated mRNA revealed enrichment of genes encoding diacylglycerol kinases, phosphatidylinositol kinases, phospholipases, integrins, and laminins.
Materials and Methods
Construction of Plasmids
A Cre cDNA originating from Type I topoisomerase of bacteriophage P1 fragment was amplified from a pAAV-Cre plasmid (AAV-401, Cell Biolabs). The nuclear localization signal (NLS) of the SV40 large T antigen sequence (5’-GAGCACCATGCCAAAAAAGAAGAGAAAGGTATCCAATTTAC-3’) was synthesized by Sigma–Aldrich. A 3519-bp mouse pColQ1a promoter (positions 31,557,371–31,553,853 on chromosome 14 according to GRCm38/mm10) was amplified by PCR using genomic DNA of C57BL/6 mouse. We ligated pColQ1a, NLS, and Cre cDNA to generate pColQ1a-Cre (Figure 1B). As a control, we generated pCMV-Cre. Primers for PCR are shown in Supplementary Table S1. Plasmids were purified with the EndoFree Megaprep DNA Purification System (Qiagen). The absence of artifacts in the plasmid inserts was confirmed by Sanger sequencing.
Immunofluorescent Staining
TA and soleus muscles were harvested and fixed in 4% paraformaldehyde (PFA) for 1 h and were teased into muscle fibers in phosphate-buffered saline (PBS). The teased fibers were incubated in 100 mM glycine in PBS for 15 min at room temperature (RT). The fibers were washed in PBS three times for 15 min each and were incubated in a blocking buffer (2% bovine serum albumin, 5% goat serum, and 0.5% Triton X-100 in PBS) for 1 h at RT. After blocking, the fibers were incubated with mouse monoclonal anti-Cre recombinase antibody (1:100, MAB3120, Millipore) or anti-HA antibody (1:200, Covance) in a blocking buffer overnight at 4°C. After washed in 0.1% Triton X-100 in PBS several times, the fibers were incubated with FITC-conjugated goat anti-mouse IgG (1:100, FI2000, Vector Laboratories). AChR was stained with Alexa 594-conjugated α-bungarotoxin (1:500, B13423, Invitrogen) for 2 h at RT. Finally, the fibers were covered by a coverslip with Vectashield with 4’,6-diamidino-2-phenylindole (DAPI; Vector Laboratories). Fluorescence signals were quantified by the IX71 microscope (Olympus) equipped with the MetaMorph imaging software (Molecular Devices).
Frozen sections of TA muscle at 10 μm were incubated with a primary antibody (anti-HA antibody at 1:200, Covance) overnight at 4°C. The sections were added with FITC-conjugated goat anti-mouse IgG (1:100, FI2000, Vector Laboratories) and Alexa 594-conjugated α-bungarotoxin (1:500, B13423, Invitrogen) followed by incubation for 1 h at RT. Finally, the sections were covered by a coverslip with Vectashield with DAPI, and fluorescence signals were quantified as stated above.
Electroporation of Plasmids Into Mouse TA and Soleus Muscle
All mouse experiments were approved by the Animal Care and Use Committee of the Nagoya University Graduate School of Medicine and were performed following the relevant guidelines. Under deep anesthesia of a 6-week-old C57BL/6 mouse, 36 U hyaluronidase (H4272, SIGMA) was injected into the TA muscle 2 h before injection of pCMV-Cre or pColQ1a-Cre. Then, 50 μl of each plasmid (1,000 ng/μl) was injected into the TA or soleus muscle and applied electrical pulses at 50 volts. The electric pulses were delivered in parallel to the muscle fibers with forceps-shaped electrodes (CUY650P3, Nepa Gene) connected to an electric pulse generator (NEPA21, Nepa Gene). The electrical pulses were comprised of three pulses of a rectangular wave for 25 ms at a rate of 3 Hz, followed by three pulses of the opposite polarity with the same protocol. TA or soleus muscle was harvested on day 5 after electroporation.
Generation of pColQ1a-Cre Mouse
A fragment of pColQ1a-Cre (Figure 1B) was microinjected into fertilized eggs of C57BL/6 mouse. The microinjection and subsequent transfer to foster mothers were technically supported by the Division of Experimental Animals, Nagoya University Graduate School of Medicine.
Generation of pColQ1a-Cre:RiboTag Mouse
RiboTag mice were kindly provided by Dr. Taiji Matsusaka at Tokai University under the permission of Dr. Paul S. Amieux at Western Washington University (Sanz et al., 2009). RiboTag mouse was mated to pColQ1a-Cre mouse to obtain pColQ1a-Cre:RiboTag mouse.
Immunoprecipitation Assays
Immunoprecipitation (IP) of pooled TA muscles of 6-week-old pColQ1a-Cre:RiboTag mice was performed as described previously with minor modifications (Sanz et al., 2009). Briefly, TA muscle was mashed by FastPrep 24 Instrument (MBP) in an IP buffer (50 mM Tris-HCl, pH 7.5, 100 mM KCl, 12 mM MgCl2, 1% Nonidet P-40, 1 mM DTT, 400 units/ml Toyobo RNase Inhibitor, 1 mg/ml heparin, 100 μg/ml cycloheximide, and the cOmplete protease inhibitor (one tablet in 25 ml, Roche). Then, anti-HA antibody (1:300, Covance) was added to 1,200 μl of cleared homogenates and incubated for 6 h at 4°C. Then protein G magnetic beads (Dynabeads, Invitrogen) in an IP buffer were added and incubated overnight at 4°C. After incubation, beads were washed four times for 5 min in a high-salt buffer (50 mM Tris-HCl, pH 7.5, 300 mM KCl, 12 mM MgCl2, 1% Nonidet P-40, 1 mM DTT, and 100 μg/ml cycloheximide) in the presence of 400 units/ml RNase Inhibitor (Toyobo) and the cOmplete™ protease inhibitor (one tablet in 25 ml, Roche). RNA was eluted in an RLT buffer with β-mercaptoethanol (Qiagen) and was purified by RNeasy Micro Kit (Qiagen) with DNase treatment. Total RNA was quantified and its quality was evaluated by RNA integrity number (RIN; Schroeder et al., 2006) representing the quality and integrity of RNA using an Agilent 2100 Bioanalyzer with the RNA 6000 Pico kit (Agilent Technologies).
Gene Expression Analysis
After confirming that the RIN was higher than 7.0, total RNA was reverse-transcribed using random hexamer primers (Thermo Fisher Scientific) and ReverTra Ace (Toyobo). Quantitative RT-PCR (qRT-PCR) was performed with the LightCycler 480 (Roche Applied Science) using the TB Green Premix ExTaq II (Takara Bio). Gene expression levels were normalized to that of glyceraldehyde-3-phosphate dehydrogenase (Gapdh), and also to the ratio of input RNA. For both IP RNA and input RNA, TA muscles of 10, 11, 12, and 16 pColQ1a-Cre:Ribotag mice were pooled into four independent samples to be analyzed in quadruplicate. qRT-PCR primers are indicated in Supplementary Table S2.
High-Throughput RNA Sequencing (RNA-Seq)
In the four pooled pairs of IP RNA and input RNA samples stated above, a pair of samples arising from 11 pColQ1a-Cre:Ribotag mice had the highest RIN in both IP RNA and input RNA, and was subjected to RNA-seq. Quality of the RNA samples was checked using both an Agilent TapeStation and an Agilent 2100 Bioanalyzer with the RNA 6000 Nano Kit, and the following thresholds were applied: quantity (>100 ng), concentration (>1 ng/μl), no contamination of DNA, no degradation of RNA confirmed by rRNA peaks (18S, 28S), and RIN (>7.0). RNA-seq of Input RNA and IP RNA was performed at Macrogen and BGI, respectively. At both companies, a sequencing library was prepared using the TruSeq Stranded mRNA kit (Illumina), and the library was read on Illumina NovaSeq 6000 (150 bp paired-end reads). As the amount of IP RNA remained low even after pooling TA muscles, IP RNA was linearly amplified by the Smart-seq2 protocols (Picelli et al., 2014). Raw reads were mapped to the mouse reference genome (NCBI37/mm9) using STAR version 2.5.3a (Dobin et al., 2013) with standard options for the long RNA-seq pipeline by ENCODE (Consortium, 2012). Transcripts per million (TPM) of each gene was calculated by RSEM version 1.3.1 with default parameters (Li and Dewey, 2011).
Gene Set Enrichment Analysis (GSEA)
We performed pathway analysis with Gene Set Enrichment Analysis (GSEA1; Vickers et al., 2008) using the following parameters: the number of permutations was 1,000; the phenotype labels were “IP vs. input;” the collapse/remap to gene symbols was “collapse;” the permutation type was configured to the “gene set” to avoid the potential problem of small sample size; and the chip platform was “Mouse Gene Symbol Remapping to Human Orthologs MSigDB.v7.1.chip.” The KEGG subset of canonical pathways (186 gene sets) was used as a reference gene set. Nominal p-value <0.05 and false-discovery rate (FDR) q-value <0.25 were used as the cut-off criteria (Reimand et al., 2019).
Results
Mouse pColQ1a Promoter Expresses Cre at the NMJ of the TA Muscle
Transcriptions of the mouse ColQ gene start from exons 1 and 1a, which are driven by pColQ1 and pColQ1a promoters, respectively (Figure 1A). A 3519-bp DNA fragment of mouse pColQ1a was amplified by PCR and was added 5′ to the nuclear localization signal (NLS) and Cre cDNA to generate a pColQ1a-Cre plasmid (Figure 1B). As the skeletal muscle fibers are comprised of multi-nucleated cells, NLS-mediated confinement of Cre in the nuclei was essential.
To confirm that pColQ1a-Cre expresses Cre specifically in the subsynaptic nuclei of TA muscle, we introduced pColQ1a-Cre in the mouse hindlimb by electroporation (Figure 1C; Murakami et al., 2003; Young and Dean, 2015). pCMV-Cre was used as a control. The TA and soleus muscles were teased into single fibers to visualize the NMJs. pCMV-Cre expressed Cre throughout TA muscle, and only 12.5% of Cre-positive areas were overlapped with AChR-positive areas. In contrast, pColQ1a-Cre expressed Cre at the NMJ of TA muscle, and 86.4% of Cre-positive areas were overlapped with AChR-positive areas (Figures 1D,E and Supplementary Table S3). These results indicate that the pColQ1a promoter drove the subsynaptic expression of Cre in the TA muscle.
Next, we generated a transgenic mouse line carrying pColQ1a-Cre. The expression of Cre was again examined in teased muscle fibers of the TA and soleus muscles, which are mostly comprised of fast-twitch and slow-twitch muscle fibers, respectively (Figure 1F). In TA muscle, 63.6% of teased muscle fibers expressed Cre at the NMJ. In contrast, in soleus muscle, the ratio was only 14.9%. Notably, 52.5% of the soleus muscle fibers did not express Cre either at the NMJ or at the extrajunctional regions (Figure 1G and Supplementary Table S4). These results indicate that the 3519-bp fragment of the mouse pColQ1a promoter region restricted the expression of Cre at the NMJs of fast-twitch muscle but not of slow-twitch muscle.
pColQ1a-Cre:Ribotag Mouse Expresses HA-Tagged Ribosomal Protein RPL22 at the NMJ of TA Muscle
We next mated our pColQ1a-Cre mouse with RiboTag mouse (Sanz et al., 2009). The generated pColQ1a-Cre:Ribotag mouse should express HA-tagged ribosomal protein RPL22 specifically at the motor endplate of fast-twitch muscle. To verify that the HA-tagged ribosomes were expressed specifically at the NMJ in pColQ1a-Cre:RiboTag mice, we immunostained the HA-tag in cross-sections of TA muscle (Figure 2A) and teased TA muscle fibers (Figure 2B). Staining for the HA-tag was mostly overlapped with α-bungarotoxin-stained AChR in cross-sections of TA muscle (Figure 2A) and teased TA muscle fibers (Figure 2B).
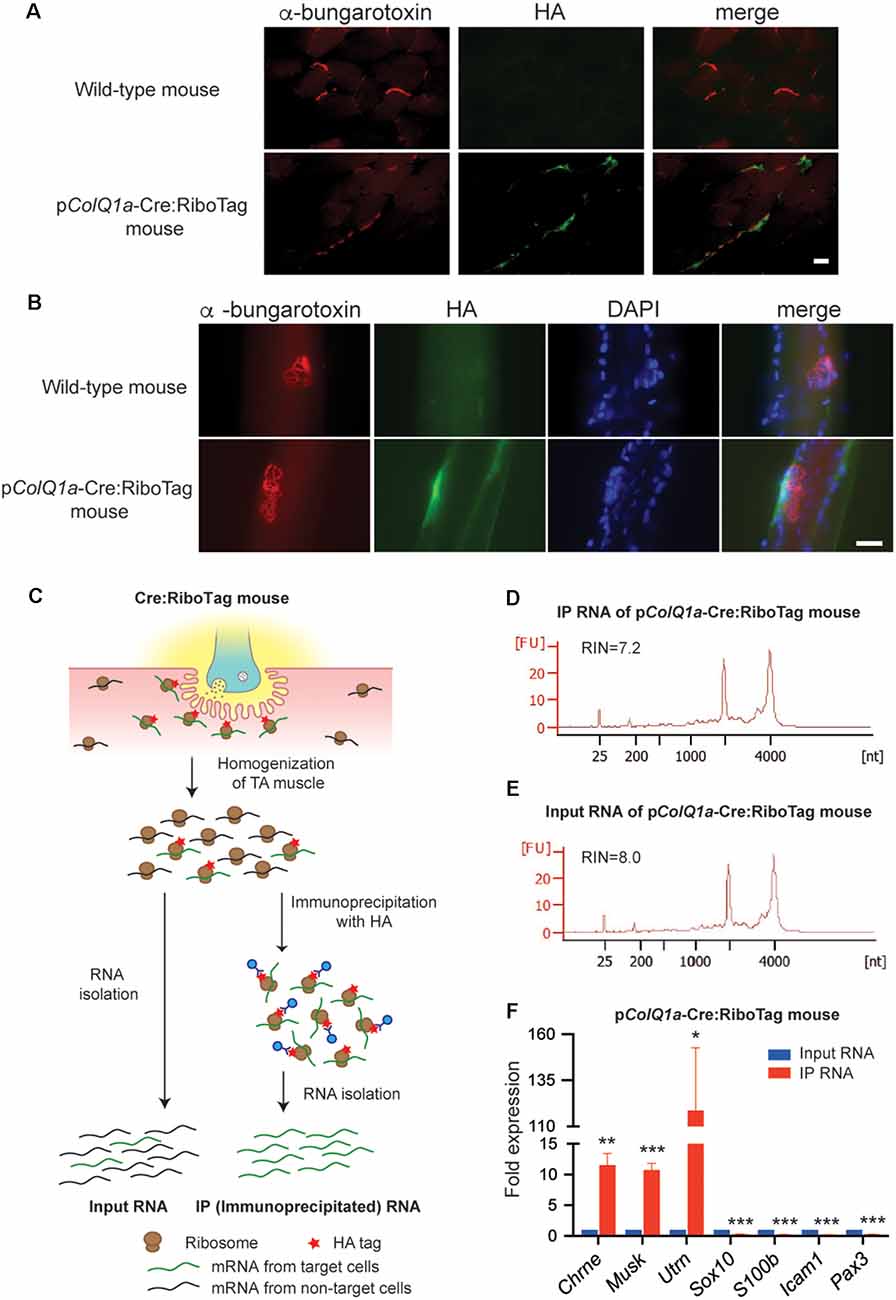
Figure 2. pColQ1a-Cre:Ribotag mouse to disclose a gene expression profile at the motor endplate TA muscle. (A,B) Immunofluorescence staining of TA muscle of wild-type and pColQ1a-Cre:RiboTag mice. Muscle sections (A) and teased muscle fibers (B) are stained with anti-HA antibody (green) and Alexa594-conjugated α-bungarotoxin 594 (red). Nuclei are stained with DAPI. Scale bar = 15 μm. (C) Schematic of the RiboTag technique for gene expression profiling at the motor endplate of TA muscle. (D,E) Agilent Technologies 2100 Bioanalyzer PicoChip outputs showing the quality of immunoprecipitated (IP) RNA (D) and input RNA (E). (F) qRT-PCR of NMJ-specific Chrne, Musk, and Utrn genes, Schwann cell-specific Sox10 and S100b, vascular endothelial cell-specific Icam1, and muscle satellite cell-specific Pax3. Gene expression level in IP RNA was normalized to that of Gapdh, and also to the ratio in input RNA. TA muscles of 10, 11, 12, and 16 pColQ1a-Cre:Ribotag mice were pooled into four independent samples, and were analyzed in quadruplicate (n = 4). Mean and SD is indicated. *p < 0.05, **p < 0.01 and ***p < 0.001 by paired Student’s t-test.
Enrichment of NMJ-Specific Genes and Paucity of Genes in Other Cell Types in TA Muscle of pColQ1a-Cre:Ribotag Mouse
To identify gene expression profiles at the motor endplate of TA muscle, we immunoprecipitated mRNA under translation using an anti-HA antibody, which was compared with input RNA (Figure 2C). We obtained ~30 ng of immunoprecipitated (IP) RNA and ~200 ng of input RNA by pooling TA muscles of 11 pColQ1a-Cre:RiboTag mice. As a control, we confirmed that only 0.67 ng RNA was immunoprecipitated by anti-HA antibody by pooling TA muscles of eight wild type mice. We confirmed that the RNA integrity numbers (RIN), representing the quality of RNA, of the IP and input RNA were more than 7.0 (Figures 2D,E). We first verified that expressions of Chrne, MuSK, and Utrn that should be concentrated in the subsynaptic nuclei were enriched in IP RNA (Figure 2F). In contrast, markers for Schwann cells (Sox10 and S100b), vascular endothelial cells (Icam1), and muscle satellite cells (Pax3) were markedly attenuated in IP RNA (Figure 2F). Therefore, IP RNA of TA muscles of pColQ1a-Cre:RiboTag mice were highly enriched in motor endplate-specific genes with scarce contamination of genes from adjacent cells at the NMJ.
Differential Gene Expression (DGE) Analysis Reveals Genes That Are Up- or Down-Regulated at the Motor Endplate of TA Muscle
To extensively characterize a gene expression profile under translation at the motor endplate of TA muscle, we performed RNA-seq analysis of IP RNA and input RNA. A scatter plot of transcripts per million (TPM) of 22,383 protein-coding genes in IP RNA and input RNA showed that the coefficient of determination (r2) was 0.6225 (Figure 3A). As have been previously applied in other studies (Srivastava et al., 2017; Wang et al., 2019), we arbitrarily filtered genes with TPM >0.5 and fold change (IP/input or input/IP) >10. Differential gene expression (DGE) analysis identified 1177 genes: 1153 genes were upregulated and 24 genes were downregulated in the subsynaptic nuclei of TA muscle. The top 20 upregulated genes and the top 20 downregulated genes are graphically shown in Figures 3B,C, respectively.
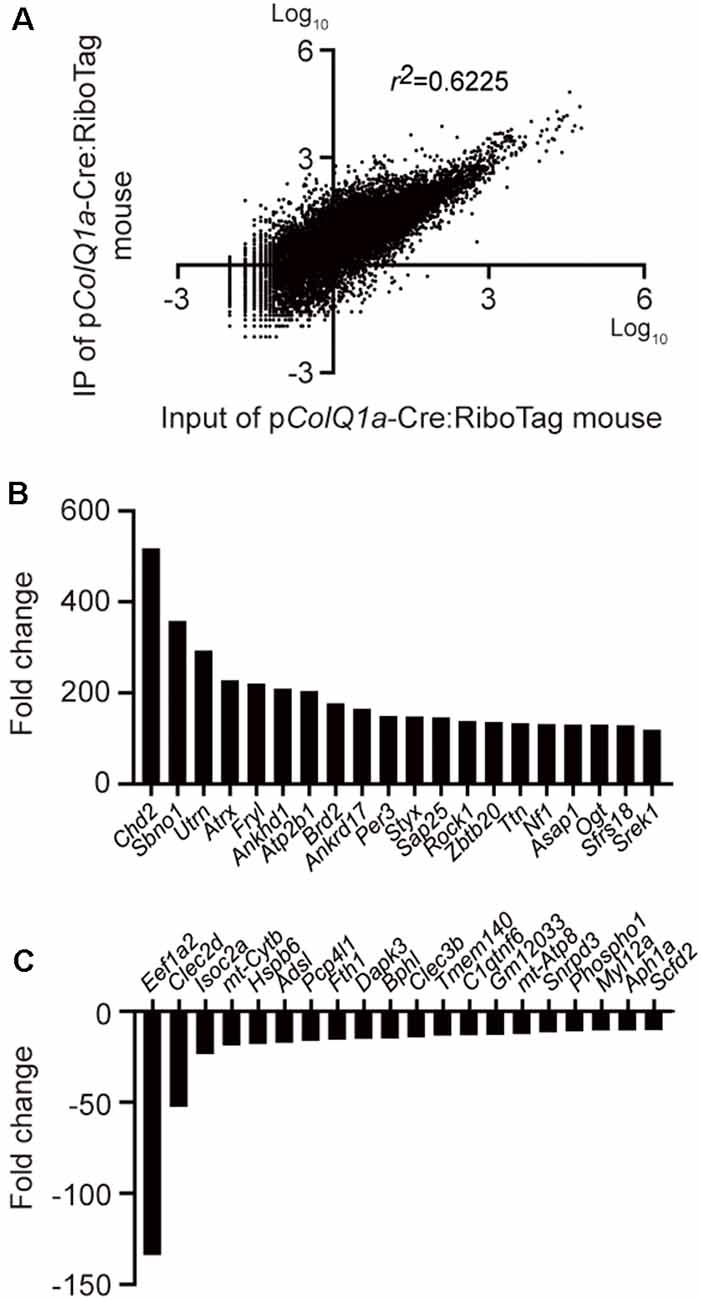
Figure 3. Differentially expressed genes between IP RNA and input RNA of the TA muscle. (A) Scatter plot of gene expressions in transcripts per million (TPM) between IP RNA and input RNA. The coefficient of determination (r2) is indicated. Representative upregulated (B) and downregulated (C) genes that were ranked top 20 each.
Gene Set Enrichment Analysis (GSEA) Reveals Key Gene Sets That Are Enriched at the Motor Endplate of TA Muscle
To further characterize gene sets that were specifically enriched at the motor endplate of TA muscle, we performed GSEA using 186 gene sets retrieved from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. We found that two gene sets of “phosphatidylinositol signaling system” and “ECM receptor interaction” were enriched at the motor endplate (nominal p < 0.05, FDR q < 0.25; Figures 4A–C and Supplementary Table S5). We also made a set of genes that are expressed at the motor endplate and are associated with myasthenia gravis and postsynaptic congenital myasthenic syndromes such as Chrne, Musk, and Dok7. GSEA analysis showed that these genes were enriched at the motor endplate of TA muscle but without statistical significance, which was likely due to the limited number of genes (p = 0.070, Supplementary Figure S1 and Supplementary Table S5). The gene set “phosphatidylinositol signaling system” included genes encoding diacylglycerol kinases (DGK*), phosphatidylinositol kinases (PIK*), and phospholipases (PL*). Similarly, the gene set “ECM receptor interaction” included genes encoding integrins (ITG*) and laminins (LAM*). We made our own gene sets for the five gene families and performed GSEA analysis, and found that these genes were indeed enriched at the motor endplate of TA muscle (Figures 4D–I and Supplementary Table S5).
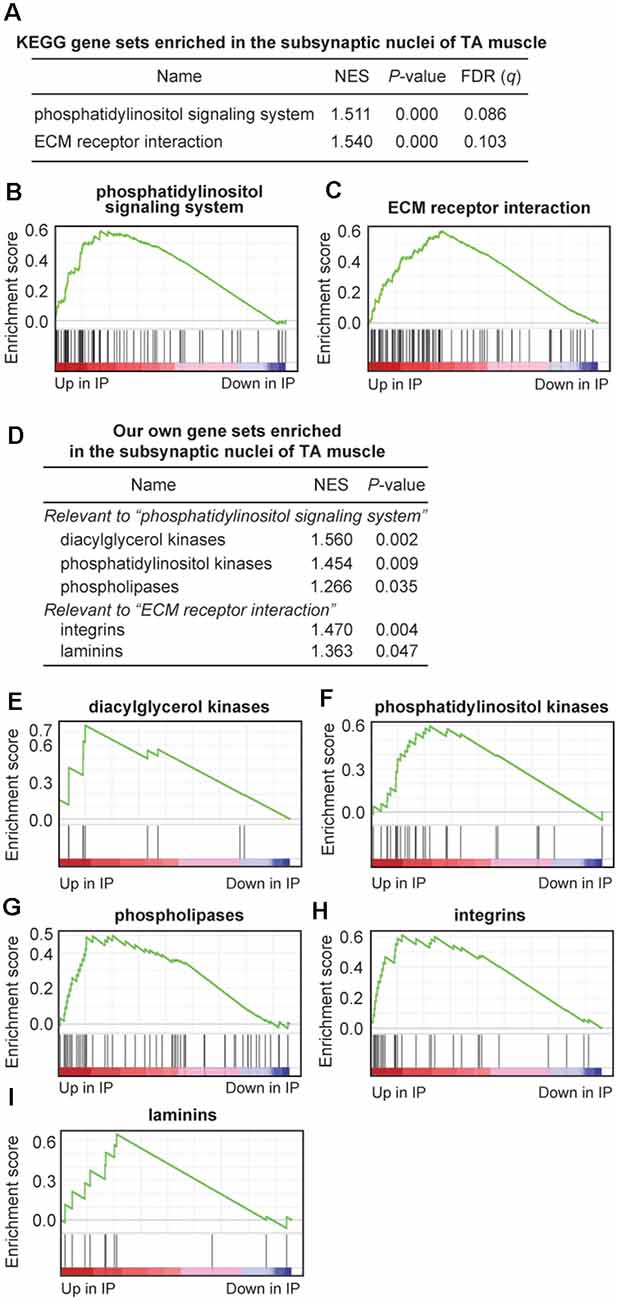
Figure 4. Gene sets enriched at the motor endplate of TA muscle by Gene Set Enrichment Analysis (GSEA) analysis. (A–C) Two KEGG gene sets were enriched at the motor endplate of TA muscle, which exceeded the thresholds of nominal p-value < 0.05 and false-discovery rate (FDR) q-value < 0.25. NES, normalized enrichment score of GSEA. (D–I) Enrichment at the motor endplate of TA muscle of our gene sets inspired by the two KEGG gene sets (A). Our own gene sets are not subsets of the two KEGG gene sets but include additional relevant genes. See Supplementary Table S5 for gene names and TPM values included in each gene set.
Discussion
We first confirmed that pColQ1a drives the expression of NLS-tagged Cre recombinase specifically at the motor endplate of TA muscle (Figure 1), and made a transgenic pColQ1a-Cre mouse line. We then mated RiboTag mouse with the pColQ1a-Cre mouse to make pColQ1a-Cre:RiboTag mouse, which expressed an HA-tagged RPL22 specifically at the motor endplate of TA muscle (Figure 2) and enabled NMJ-specific gene expression profiling. mRNA under translation at the motor endplate was co-immunoprecipitated by an anti-HA antibody (Figure 2C). We confirmed that motor endplate-specific Chrne and Musk were enriched in IP RNA, whereas Schwann cell-specific Sox10 and S100b, vascular endothelial cell-specific Icam1, and muscle satellite cell-specific Pax3 were much reduced in IP RNA (Figure 2F). We then analyzed IP RNA by RNA-seq. We compared IP RNA with input RNA obtained from whole TA muscle to characterize motor endplate-specific gene expression profile of TA muscle (Figures 3, 4).
The mechanisms of NMJ-specific transcriptions of ColQ has been previously reported. Laser capture microdissection of the mouse NMJ revealed NMJ-specific expression of ColQ in the extraocular muscles (Ketterer et al., 2010) and the TA muscle (Nazarian et al., 2005; Ketterer et al., 2010). The NMJ-specific expression of ColQ is mediated by transcriptional upregulation at the NMJ and transcriptional downregulation in the extra-junctional regions. The E-box (CANNTG) in pColQ1a binds to the myogenic differentiation factors (MDFs) of the MyoD family and drives muscle-specific gene expression (Lee et al., 2004). Contrarily, muscle action potentials suppress the E-box at the extra-junctional regions to drive NMJ-specific expression of an E-box-bearing gene (Ting et al., 2005). Transcription of most muscle-specific genes, however, are not suppressed by electrical activity, and other unidentified cis-elements are likely to be operational to achieve electrical transcriptional suppression (Ohno et al., 1999). Transcriptional upregulation of pColQ1a at the NMJ is mediated by the N-box (CCGGAA; Lee et al., 2004). The N-box (CGGAA) is also the neuregulin-response element in mouse Chrnd encoding the δ subunit of acetylcholine receptor (AChR; Fromm and Burden, 1998) and in rat Chrne encoding the AChR ε subunit (Sapru et al., 1998). Similar involvement of the neuregulin-responsive N-box in the NMJ-specific transcription of ColQ in mouse have also been reported (Lee et al., 2004; Ting et al., 2005). Also, specific activation of pColQ1 and pColQ1a in slow- and fast-twitch muscles, respectively, is mediated by a slow upstream regulatory element (SURE) in pColQ1 and a fast intronic regulatory element (FIRE) in pColQ1a (Lee et al., 2004; Ting et al., 2005).
Cell-type-specific expression analysis has gained popularity in the past decades and is of particular importance to understand the biology of certain cell types or structures. The NMJ is one such complicated example consisting of presynaptic, synaptic, and postsynaptic components within a complex tissue environment (Slater, 2017; Huang et al., 2019). As stated above, NMJ-specific gene expression profiles have been analyzed by laser capture microdissection of extraocular muscles (Ketterer et al., 2010) and TA muscle (Nazarian et al., 2005; Ketterer et al., 2010), which, however, were inevitably contaminated with vascular endothelial cells, lymphocytes, and terminal Schwann cells. pColQ1a-Cre:RiboTag mouse enabled extensive profiling of gene expressions under translation at the motor endplate of TA muscle with minimal contamination of adjacent cells. Gene expression profiling with RiboTag mouse inherently relies on the tissue- or cell-type-specific expression of HA-tagged RPL22 (Sanz et al., 2009). Although pColQ1a was likely to be specifically active at the motor endplate of TA muscle, the transgenic pColQ1a-Cre mouse carried a 3519-bp fragment of the ColQ1a promoter, which should have allowed a small amount of leaky expression of Cre recombinase at extra-junctional regions. A combination of laser capture microdissection and RiboTag may elucidate more precise gene expression profiles of the NMJ.
GSEA analysis revealed that two gene sets of “phosphatidylinositol signaling system” and “ECM receptor interaction” in the KEGG database were enriched at the subsynaptic nuclei of TA muscle (Figures 4A–C). Inspection of the two upregulated gene sets prompted us to make our own gene sets that covered more relevant genes than the KEGG database. Being prompted by the gene set of “phosphatidylinositol signaling system,” we found that genes encoding diacylglycerol kinases, phosphatidylinositol kinases, and phospholipases were enriched at the motor endplate of TA muscle (Figures 4D–G). Diacylglycerol kinases (DGK) are enzymes that phosphorylate diacylglycerol to yield phosphatidic acid. In the adult mouse, DGK-ζ encoded by DGKZ binds to syntrophin on the sarcolemma and is concentrated at the NMJ in a dystrophin-independent manner (Abramovici et al., 2003). DGK-ζ is likely to control lipid-signaling pathways to regulate actin organization (Abramovici et al., 2003). In C. elegans, presynaptic diacylglycerol facilitates the release of acetylcholine at NMJ, and DGK possibly regulates the abundance of presynaptic diacylglycerol (Lackner et al., 1999). We showed that DGK-η (DGKH) and DGK-θ (DGKQ) were 140-fold and 27-fold enriched at the motor endplate, respectively, and were more enriched than DGK-ζ (DGKZ; Supplementary Table S5). These and other DGKs may have undisclosed roles at the motor endplate. Phospholipases are enzymes that hydrolyze phospholipids into inositol phosphates and diacylglycerol, which are implicated as second messengers for the subsequent cellular events (Kadamur and Ross, 2013). Phospholipase C (PLC), one of the major classes of phospholipases, evokes endplate responses to acetylcholine in rodent skeletal muscle (Harborne et al., 1987). Two other classes of phospholipases are likely to have presynaptic roles. Phospholipase D blocks acetylcholine release by reducing the number of active presynaptic-releasing sites (Humeau et al., 2001). Phospholipase A2 regulates acetylcholine release with an inhibitory or excitatory effect in several distinct phases by controlling the Ca2+ or K+ channels (Fathi et al., 2001). We showed that phospholipases D and A2 are also enriched at the motor endplate (Supplementary Table S5), and these phospholipases may exert their effects by signaling through the motor endplate to the nerve terminal or by exerting undisclosed roles at the motor endplate. Similarly, by being prompted by the gene set of “ECM receptor interaction,” we found that genes encoding integrins and laminins were enriched at the motor endplate of TA muscle (Figures 4D,H,I). Integrins are cell surface receptors that facilitate adhesion of ECM molecules. The roles of integrins at the NMJ have been analyzed for integrins α3 and α7β1. Integrin α3 regulates multiple aspects of the development, function, and maintenance of the NMJ (Ross et al., 2017). Integrin α3 is required for fully efficient neurotransmission at the NMJ and also for the connection of motor neurons to the muscle fibers (Ross et al., 2017). Integrin α7β1 is concentrated at the postsynaptic NMJ in human and mouse, and their abundance is slightly increased in mdx mice in response to dystrophin deficiency (Hodges et al., 1997). We showed that yet uncharacterized integrins α2b and α10 were 163-fold and 68-fold enriched at the motor endplate (Supplementary Table S5). These and other integrins may have essential effects on the NMJ. Some laminins are also enriched at the NMJ (Rogers and Nishimune, 2017). Laminins α5 and β2 are enriched at the synaptic basal lamina, and laminin α4 is surrounding the nerve terminal, whereas laminins α2, β1, and γ1 are ubiquitously expressed on muscle fibers (Nishimune et al., 2008; Singhal and Martin, 2011). We found that laminins α3 and γ3 were 25-fold and 6-fold enriched at the motor endplate, respectively (Supplementary Table S5), and these laminins may have yet undisclosed roles at the NMJ.
To summarize, we have determined a gene expression profile at the motor endplate of TA muscle, which was expected to be minimally contaminated with adjacent cells at the NMJ. We expect that our gene expression profiling will contribute to the understanding of the molecular mechanisms of development, maintenance, and pathology of the NMJ.
Data Availability Statement
FASTQ files are available at the DNA Data Bank of Japan (DDBJ) under the accession number DRA010277 (https://www.ncbi.nlm.nih.gov/sra/?term=DRA010277).
Ethics Statement
The animal study was reviewed and approved by Animal Care and Use Committee of the Nagoya University Graduate School of Medicine.
Author Contributions
KO conceived the project. KH, MI, and KO designed experiments. KH and JL performed most of the experiments with the help of MI, BO, and AM. RNA-seq analysis was performed by J-IT and TO. KH, MI, and KO wrote the article.
Funding
This study was supported by grants-in-aid by Japan Society for the Promotion of Science (18K14684, JP18K06058, JP18K06483, 19K22802, JP20H03561, and JP20K06925); Ministry of Health, Labour, and Welfare, Japan (20FC1036); Japan Agency for Medical Research and Development (19ek0109230, 20gm1010002, 20ek0109281, and 20bm080400); and National Center of Neurology and Psychiatry (2-5).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge Dr. Taiji Matsusaka at Tokai University and Dr. Paul S. Amieux at Western Washington University for providing us with RiboTag mouse. We also acknowledge the staff at the Division of Experimental Animals, Nagoya University Graduate School of Medicine for their technical support. We would like to thank Ms. Keiko Itano at Nagoya University for creating artworks.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2020.00154/full#supplementary-material.
FIGURE S1 | GSEA analysis of a set of genes that are associated with myasthenia gravis and postsynaptic congenital myasthenic syndromes. See Supplementary Table S5 for gene names and TPM values included in the gene set.
TABLE S1 | Primer sequences for plasmid construction and genotyping.
TABLE S2 | Primer sequences for qRT-PCR.
TABLE S3 | The Numbers of Cre-positive and Cre-negative NMJs in teased TA muscle fibers electroporated with pCMV-Cre or pColQ1a-Cre.
TABLE S4 | Distribution of Cre-positive signals in teased muscle fibers of the TA and soleus muscles of the pColQ1a-Cre mouse.
TABLE S5 | Detailed values of RNA-seq analysis. TPM of 22,382 genes in input RNA and IP RNA. Results of GSEA analysis of two KEGG gene sets and five our own gene sets that were enriched at the motor endplate of TA muscle.
References
Abramovici, H., Hogan, A. B., Obagi, C., Topham, M. K., and Gee, S. H. (2003). Diacylglycerol kinase-zeta localization in skeletal muscle is regulated by phosphorylation and interaction with syntrophins. Mol. Biol. Cell. 14, 4499–4511. doi: 10.1091/mbc.e03-03-0190
Consortium, E. P. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. doi: 10.1038/nature11247
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Fathi, H. B., Rowan, E. G., and Harvey, A. L. (2001). The facilitatory actions of snake venom phospholipase A(2) neurotoxins at the neuromuscular junction are not mediated through voltage-gated K(+) channels. Toxicon 39, 1871–1882. doi: 10.1016/s0041-0101(01)00170-2
Fromm, L., and Burden, S. J. (1998). Synapse-specific and neuregulin-induced transcription require an ets site that binds GABPα/GABPβ. Genes Dev. 12, 3074–3083. doi: 10.1101/gad.12.19.3074
Gaspersic, R., Koritnik, B., Crne-Finderle, N., and Sketelj, J. (1999). Acetylcholinesterase in the neuromuscular junction. Chem. Biol. Interact. 119-120, 301–308. doi: 10.1016/s0009-2797(99)00040-x
Harborne, A. J., Turner, A. W., Murphy, R. L., and Smith, M. E. (1987). Action of phospholipase C at the neuromuscular junction in rodent skeletal muscles. Eur. J. Pharmacol. 143, 163–170. doi: 10.1016/0014-2999(87)90529-2
Hirsch, N. P. (2007). Neuromuscular junction in health and disease. Br. J. Anaesth. 99, 132–138. doi: 10.1093/bja/aem144
Hodges, B. L., Hayashi, Y. K., Nonaka, I., Wang, W., Arahata, K., and Kaufman, S. J. (1997). Altered expression of the α7β1 integrin in human and murine muscular dystrophies. J. Cell. Sci. 110, 2873–2881.
Huang, K., Luo, Y. B., and Yang, H. (2019). Autoimmune channelopathies at neuromuscular junction. Front. Neurol. 10:516. doi: 10.3389/fneur.2019.00516
Humeau, Y., Vitale, N., Chasserot-Golaz, S., Dupont, J. L., Du, G., Frohman, M. A., et al. (2001). A role for phospholipase D1 in neurotransmitter release. Proc. Natl. Acad. Sci. U S A 98, 15300–15305. doi: 10.1073/pnas.261358698
Kadamur, G., and Ross, E. M. (2013). Mammalian phospholipase C. Annu. Rev. Physiol. 75, 127–154. doi: 10.1146/annurev-physiol-030212-183750
Ketterer, C., Zeiger, U., Budak, M. T., Rubinstein, N. A., and Khurana, T. S. (2010). Identification of the neuromuscular junction transcriptome of extraocular muscle by laser capture microdissection. Invest. Ophthalmol. Vis. Sci. 51, 4589–4599. doi: 10.1167/iovs.09-4893
Kim, T., Lim, C. S., and Kaang, B. K. (2015). Cell type-specific gene expression profiling in brain tissue: comparison between TRAP, LCM and RNA-seq. BMB Rep. 48, 388–394. doi: 10.5483/bmbrep.2015.48.7.218
Krejci, E., Legay, C., Thomine, S., Sketelj, J., and Massoulie, J. (1999). Differences in expression of acetylcholinesterase and collagen Q control the distribution and oligomerization of the collagen-tailed forms in fast and slow muscles. J. Neurosci. 19, 10672–10679. doi: 10.1523/jneurosci.19-24-10672.1999
Lackner, M. R., Nurrish, S. J., and Kaplan, J. M. (1999). Facilitation of synaptic transmission by EGL-30 Gqα and EGL-8 PLCβ: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron 24, 335–346. doi: 10.1016/s0896-6273(00)80848-x
Lee, H. H., Choi, R. C., Ting, A. K., Siow, N. L., Jiang, J. X., Massoulie, J., et al. (2004). Transcriptional regulation of acetylcholinesterase-associated collagen ColQ: differential expression in fast and slow twitch muscle fibers is driven by distinct promoters. J. Biol. Chem. 279, 27098–27107. doi: 10.1074/jbc.m402596200
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Murakami, T., Nishi, T., Kimura, E., Goto, T., Maeda, Y., Ushio, Y., et al. (2003). Full-length dystrophin cDNA transfer into skeletal muscle of adult mdx mice by electroporation. Muscle Nerve 27, 237–241. doi: 10.1002/mus.10283
Nazarian, J., Bouri, K., and Hoffman, E. P. (2005). Intracellular expression profiling by laser capture microdissection: three novel components of the neuromuscular junction. Physiol. Genomics 21, 70–80. doi: 10.1152/physiolgenomics.00227.2004
Nazim, M., Masuda, A., Rahman, M. A., Nasrin, F., Takeda, J. I., Ohe, K., et al. (2017). Competitive regulation of alternative splicing and alternative polyadenylation by hnRNP H and CstF64 determines acetylcholinesterase isoforms. Nucleic Acids Res. 45, 1455–1468. doi: 10.1093/nar/gkw823
Nishimune, H., Valdez, G., Jarad, G., Moulson, C. L., Muller, U., Miner, J. H., et al. (2008). Laminins promote postsynaptic maturation by an autocrine mechanism at the neuromuscular junction. J. Cell. Biol. 182, 1201–1215. doi: 10.1083/jcb.200805095
Ohno, K., Anlar, B., and Engel, A. G. (1999). Congenital myasthenic syndrome caused by a mutation in the Ets-binding site of the promoter region of the acetylcholine receptor epsilon subunit gene. Neuromuscul. Disord. 9, 131–135. doi: 10.1016/s0960-8966(99)00007-3
Ohno, K., Brengman, J., Tsujino, A., and Engel, A. G. (1998). Human endplate acetylcholinesterase deficiency caused by mutations in the collagen-like tail subunit (ColQ) of the asymmetric enzyme. Proc. Natl. Acad. Sci. U S A 95, 9654–9659. doi: 10.1073/pnas.95.16.9654
Ohno, K., Ohkawara, B., and Ito, M. (2017). Agrin-LRP4-MuSK signaling as a therapeutic target for myasthenia gravis and other neuromuscular disorders. Expert Opin. Ther. Targets 21, 949–958. doi: 10.1080/14728222.2017.1369960
Picelli, S., Faridani, O. R., Bjorklund, A. K., Winberg, G., Sagasser, S., and Sandberg, R. (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181. doi: 10.1038/nprot.2014.006
Reimand, J., Isserlin, R., Voisin, V., Kucera, M., Tannus-Lopes, C., Rostamianfar, A., et al. (2019). Pathway enrichment analysis and visualization of omics data using g:profiler, GSEA, cytoscape and enrichmentmap. Nat. Protoc. 14, 482–517. doi: 10.1038/s41596-018-0103-9
Rogers, R. S., and Nishimune, H. (2017). The role of laminins in the organization and function of neuromuscular junctions. Matrix Biol. 57-58, 86–105. doi: 10.1016/j.matbio.2016.08.008
Ross, J. A., Webster, R. G., Lechertier, T., Reynolds, L. E., Turmaine, M., Bencze, M., et al. (2017). Multiple roles of integrin-α3 at the neuromuscular junction. J. Cell. Sci. 130, 1772–1784. doi: 10.1242/jcs.201103
Sanz, E., Yang, L., Su, T., Morris, D. R., Mcknight, G. S., and Amieux, P. S. (2009). Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. U S A 106, 13939–13944. doi: 10.1073/pnas.0907143106
Sapru, M. K., Florance, S. K., Kirk, C., and Goldman, D. (1998). Identification of a neuregulin and protein-tyrosine phosphatase response element in the nicotinic acetylcholine receptor epsilon subunit gene: regulatory role of an Rts transcription factor. Proc. Natl. Acad. Sci. U S A 95, 1289–1294. doi: 10.1073/pnas.95.3.1289
Schroeder, A., Mueller, O., Stocker, S., Salowsky, R., Leiber, M., Gassmann, M., et al. (2006). The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7:3. doi: 10.1186/1471-2199-7-3
Singhal, N., and Martin, P. T. (2011). Role of extracellular matrix proteins and their receptors in the development of the vertebrate neuromuscular junction. Dev. Neurobiol. 71, 982–1005. doi: 10.1002/dneu.20953
Slater, C. R. (2017). The structure of human neuromuscular junctions: some unanswered molecular questions. Int. J. Mol. Sci. 18:2183. doi: 10.3390/ijms18102183
Srivastava, R., Budak, G., Dash, S., Lachke, S. A., and Janga, S. C. (2017). Transcriptome analysis of developing lens reveals abundance of novel transcripts and extensive splicing alterations. Sci. Rep. 7:11572. doi: 10.1038/s41598-017-10615-4
Ting, A. K., Siow, N. L., Kong, L. W., and Tsim, K. W. (2005). Transcriptional regulation of acetylcholinesterase-associated collagen ColQ in fast- and slow-twitch muscle fibers. Chem. Biol. Interact. 157-158, 63–70. doi: 10.1016/j.cbi.2005.10.009
Vickers, A. J., Cronin, A. M., Elkin, E. B., and Gonen, M. (2008). Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med. Inform. Decis. Mak. 8:53. doi: 10.1186/1472-6947-8-53
Wang, K., Wang, D., Zheng, X., Qin, A., Zhou, J., Guo, B., et al. (2019). Multi-strategic RNA-seq analysis reveals a high-resolution transcriptional landscape in cotton. Nat. Commun. 10:4714. doi: 10.1038/s41467-019-12575-x
Keywords: neuromuscular junction, motor endplate, collagen Q, ribotag mouse, gene expression profile
Citation: Huang K, Li J, Ito M, Takeda J-I, Ohkawara B, Ogi T, Masuda A and Ohno K (2020) Gene Expression Profile at the Motor Endplate of the Neuromuscular Junction of Fast-Twitch Muscle. Front. Mol. Neurosci. 13:154. doi: 10.3389/fnmol.2020.00154
Received: 30 May 2020; Accepted: 27 July 2020;
Published: 08 September 2020.
Edited by:
Ruth Herbst, Medical University of Vienna, AustriaReviewed by:
Said Hashemolhosseini, University of Erlangen Nuremberg, GermanyKarl Tsim, Hong Kong University of Science and Technology, Hong Kong
Copyright © 2020 Huang, Li, Ito, Takeda, Ohkawara, Ogi, Masuda and Ohno. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kinji Ohno, ohnok@med.nagoya-u.ac.jp
 Kun Huang
Kun Huang Jin Li1
Jin Li1  Bisei Ohkawara
Bisei Ohkawara Akio Masuda
Akio Masuda Kinji Ohno
Kinji Ohno