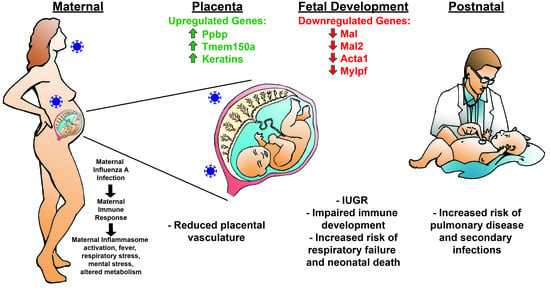
Maternal Influenza A Virus Infection Restricts Fetal and Placental Growth and Adversely Affects the Fetal Thymic Transcriptome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Influenza Virus Inoculum Preparation and Titration
2.3. Experimental Design and Sample Collection
2.4. Histopathology
2.5. Hemagglutination Inhibition (HI) Assay
2.6. Fetal Sex Determination
2.7. RNA Extraction
2.8. RNA Seq of E7.5 and E12.5 Placentae and E7.5 Thymus Pools
2.9. Pathway Analysis
2.10. Influenza A Virus Quantitative Real-Time Polymerase Chain Reaction
2.11. RT qPCR of Placentae and Thymus Pools
2.11.1. Primer Design
2.11.2. Target Gene Selection
2.11.3. Real-Time Polymerase Chain Reaction (RT-PCR) and RT-qPCR
2.12. Statistical Analysis
3. Results
3.1. Maternal Influenza Infection Induces Fetal Growth Restriction and Decreased Placental Weight
3.2. Placental Transcriptome and Histopathology in Influenza-Inoculated and Control Litters
3.3. Maternal Influenza Infection at E7.5 Is Associated with Transcript Downregulation of 957 Genes in the Fetal Thymus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barker, D.J.P. Fetal and infant origins of adult disease. Br. Med. J. 1992, 301, 1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almond, D. Is the 1918 influenza pandemic over? Long-term effects of in utero influenza exposure in the post-1940 US population. J. Polit. Econ. 2006, 114, 672–712. [Google Scholar] [CrossRef] [Green Version]
- Neelsen, S.; Stratmann, T. Long-run effects of fetal influenza exposure: Evidence from Switzerland. Soc. Sci. Med. 2012, 74, 58–66. [Google Scholar] [CrossRef]
- Doyle, T.J.; Goodin, K.; Hamilton, J.J. Maternal and neonatal outcomes among pregnant women with 2009 pandemic influenza A(H1N1) illness in Florida, 2009–2010: A population-based cohort study. PLoS ONE 2013, 8, e79040. [Google Scholar] [CrossRef] [PubMed]
- Luteijn, J.M.; Brown, M.J.; Dolk, H. Influenza and congenital anomalies: A systematic review and meta-analysis. Hum. Reprod. 2014, 29, 809–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newsome, K.W.J.; Way, S.; Honein, M.; Hill, H.; Rasmussen, S.; McIntyre, A.F.; Finelli, L.; Jamieson, C.W.; Zotti, M. Maternal and infant outcomes among severely ill pregnant and postpartum women with 2009 pandemic influenza A (H1N1)—United States, April 2009–August 2010. Morb. Mortal. Wkly. Rep. 2011, 60, 1193–1196. [Google Scholar]
- Brown, A.S.; Derkits, E.J. Prenatal infection and schizophrenia: A review of epidemiologic and translational studies. Am. J. Psychiatry 2010, 167, 261–280. [Google Scholar] [CrossRef] [Green Version]
- Khandaker, G.; Zimbron, J.; Lewis, G.; Jones, P.B. Prenatal maternal infection, neurodevelopment and adult schizophrenia: A systematic review of population-based studies. Psychol. Med. 2013, 43, 239–257. [Google Scholar] [CrossRef] [Green Version]
- Oster, M.E.; Riehle-Colarusso, T.; Alverson, C.J.; Correa, A. Associations between maternal fever and influenza and congenital heart defects. J. Pediatr. 2011, 158, 990–995. [Google Scholar] [CrossRef]
- Csaky-Szunyogh, M.; Vereczkey, A.; Kosa, Z.; Urban, R.; Czeizel, A.E. Association of maternal diseases during pregnancy with the risk of single ventricular septal defects in the offspring—A population-based case-control study. J. Matern. Fetal Neonatal Med. 2013, 26, 738–747. [Google Scholar] [CrossRef]
- Li, M.; Liu, Z.; Lin, Y.; Chen, X.; Li, S.; You, F.; Deng, Y.; Li, N.; Wang, Y.; Zhang, Y.; et al. Maternal influenza-like illness, medication use during pregnancy and risk of congenital heart defects in offspring. J. Matern. Fetal Neonatal Med. 2014, 27, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, B.A.D.; Park, K. Crimmins EM and CE Finch, Lingering prenatal effects of the 1918 influenza pandemic on cardiovascular disease. J. Dev. Orig. Health Dis. 2010, 1, 26–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, D.W.; Kim, K.W.; Rawlinson, W.D.; Craig, M.E. Maternal virus infections in pregnancy and type 1 diabetes in their offspring: Systematic review and meta-analysis of observational studies. Rev. Med. Virol. 2018, 28, e1974. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Zhang, L.; Qu, Y.; Mu, D. Meta-analysis of antenatal infection and risk of asthma and eczema. Med. Baltim. 2016, 95, e4671. [Google Scholar] [CrossRef]
- Marcelin, G.; Aldridge, J.R.; Duan, S.; Ghoneim, H.E.; Rehg, J.; Henju, M.; Boon, A.C.M.; McCullers, J.A.; Webby, R.J. Fatal Outcome of Pandemic H1N1 2009 Influenza Virus Infection Is Associated with Immunopathology and Impaired Lung Repair, Not Enhanced Viral Burden, in Pregnant Mice. J. Virol. 2011, 85, 11208–11219. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Fatemi, S.H.; Sidwell, R.W.; Patterson, P.H. Maternal influenza infection causes marked behavioral and pharmacological changes in the offspring. J. Neurosci. 2003, 23, 297–302. [Google Scholar] [CrossRef]
- Fatemi, S.H.; Folsom, T.D.; Reutiman, T.J.; Sidwell, R.W. Viral regulation of aquaporin 4, connexin 43, microcephalin and nucleolin. Schizophr. Res. 2008, 98, 163–177. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.H.; Zhang, A.J.; To, K.K.; Chan, C.C.; Poon, V.K.; Guo, K.; Ng, F.; Zhang, Q.W.; Leung, V.H.; Cheung, A.N.; et al. Wild type and mutant 2009 pandemic influenza A (H1N1) viruses cause more severe disease and higher mortality in pregnant BALB/c mice. PLoS ONE 2010, 5, e13757. [Google Scholar] [CrossRef] [Green Version]
- Littauer, E.Q.; Esser, E.S.; Antao, O.Q.; Vassilieva, E.V.; Compans, R.W.; Skountzou, I. H1N1 influenza virus infection results in adverse pregnancy outcomes by disrupting tissue-specific hormonal regulation. PLoS Pathog. 2017, 13, e1006757. [Google Scholar] [CrossRef] [Green Version]
- Williams, K.; Mackenzie, J.S. Influenza infections during pregnancy in the mouse. J. Hyg. Lond. 1977, 79, 249–257. [Google Scholar] [CrossRef] [Green Version]
- Fatemi, S.H.; Folsom, T.D.; Rooney, R.J.; Mori, S.; Kornfield, T.E.; Reutiman, T.J.; Kneeland, R.E.; Liesch, S.B.; Hua, K.G.; Hsu, J.; et al. The viral theory of schizophrenia revisited: Abnormal placental gene expression and structural changes with lack of evidence for H1N1 viral presence in placentae of infected mice or brains of exposed offspring. Neuropharmacology 2012, 62, 1290–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobets, N.V.; Deeva, A.V.; Mirchink, E.P.; Pronin, A.P.; Chernyakhovskaya, I.Y.; Zuev, V.A. The Possible Immunological Mechanisms of Pathology in Mice with Congenitally Acquired Influenza Infection. Russ. J. Immunol. 1997, 2, 121–128. [Google Scholar] [PubMed]
- Jaïdane, H.; Halouani, A.; Jmii, H.; Elmastour, F.; Abdelkefi, S.; Bodart, G.; Michaux, H.; Chakroun, T.; Sane, F.; Mokni, M. In-utero coxsackievirus B4 infection of the mouse thymus. Clin. Exp. Immunol. 2017, 187, 399–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georges, H.M.; Knapek, K.J.; Bielefeldt-Ohmann, H.; Van Campen, H.; Hansen, T.R. Attenuated lymphocyte activation leads to the development of immunotolerance in bovine fetuses persistently infected with BVDV. Biol. Reprod. 2020, 103, 560–571. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, N.P.; Webb, B.T.; McGill, J.L.; Schaut, R.G.; Bielefeldt-Ohmann, H.; Van Campen, H.; Sacco, R.E.; Hansen, T.R. Induction of interferon-gamma and downstream pathways during establishment of fetal persistent infection with bovine viral diarrhea virus. Virus Res. 2014, 183C, 95–106. [Google Scholar] [CrossRef]
- Knapek, K.J.; Georges, H.M.; Van Campen, H.; Bishop, J.V.; Bielefeldt-Ohmann, H.; Smirnova, N.P.; Hansen, T.R. Fetal Lymphoid Organ Immune Responses to Transient and Persistent Infection with Bovine Viral Diarrhea Virus. Viruses 2020, 12, 816. [Google Scholar] [CrossRef]
- Muñoz-Zanzi, C.A.; Hietala, S.K.; Thurmond, M.C.; Johnson, W.O. Quantification, risk factors, and health impact of natural congenital infection with bovine viral diarrhea virus in dairy calves. Am. J. Vet. Res. 2003, 64, 358–365. [Google Scholar] [CrossRef]
- Waldner, C.L.; Kennedy, R.I. Associations between health and productivity in cow-calf beef herds and persistent infection with bovine viral diarrhea virus, antibodies against bovine viral diarrhea virus, or antibodies against infectious bovine rhinotracheitis virus in calves. Am. J. Vet. Res. 2008, 69, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Rossant, J.; Cross, J.C. Placental development: Lessons from mouse mutants. Nat. Rev. Genet. 2001, 2, 538–548. [Google Scholar] [CrossRef]
- Novak, C.M.; Lee, J.Y.; Ozen, M.; Tsimis, M.E.; Kucirka, L.M.; McLane, M.W.; Xie, L.; Kelleher, M.; Xie, H.; Jia, B.; et al. Increased placental T cell trafficking results in adverse neurobehavioral outcomes in offspring exposed to sub-chronic maternal inflammation. Brain Behav. Immun. 2019, 75, 129–136. [Google Scholar] [CrossRef]
- Bielefeldt-Ohmann, H.; Smirnova, N.P.; Tolnay, A.E.; Webb, B.T.; Antoniazzi, A.Q.; van Campen, H.; Hansen, T.R. Neuro-invasion by a ‘Trojan Horse’ strategy and vasculopathy during intrauterine flavivirus infection. Int. J. Exp. Pathol. 2012, 93, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Isaac, S.M.; Langford, M.B.; Simmons, D.G.; Adamson, S.L. Anatomy of the mouse placenta throughout gestation. In The Guide to Investigation of Mouse Pregnancy; Croy, B.A., Yamada, A.T., DeMayo, F.J., Adamson, S.L., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 69–73. [Google Scholar]
- World Health Organization. Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza; World Health Organization: Geneva, Switzerland, 2011; Available online: http://whqlibdoc.who.int/publications/2011/9789241548090_eng.pdf (accessed on 4 September 2020).
- Capel, B.; Albrecht, K.H.; Washburn, L.L.; Eicher, E.M. Migration of mesonephric cells into the mammalian gonad depends on Sry. Mech. Dev. 1999, 84, 127–131. [Google Scholar] [CrossRef]
- Spackman, E.; Senne, D.A.; Bulaga, L.L.; Myers, T.J.; Perdue, M.L.; Garber, L.P.; Lohman, K.; Daum, L.T.; Suarez, D.L. Development of Real-Time RT-PCR for the Detection of Avian Influenza Virus. Avian Dis. 2003, 47, 1079–1082. [Google Scholar] [CrossRef] [PubMed]
- Petersen, E.E.; Staples, J.E.; Meaney-Delman, D.; Fischer, M.; Ellington, S.R.; Callaghan, W.M.; Jamieson, D.J. Interim Guidelines for Pregnant Women During a Zika Virus Outbreak—United States, 2016. Morb. Mortal. Wkly. Rep. 2016, 65, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L. Congenital Viral Infection: Traversing the Uterine-Placental Interface. Annu. Rev. Virol. 2018, 5, 273–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Racicot, K.; Mor, G. Risks associated with viral infections during pregnancy. J. Clin. Investig. 2017, 127, 1591–1599. [Google Scholar] [CrossRef] [Green Version]
- Meijer, W.J.; Wensing, A.M.; Bruinse, H.W.; Nikkels, P.G. High rate of chronic villitis in placentas of pregnancies complicated by influenza A/H1N1 infection. Infect. Dis. Obs. Gynecol. 2014, 2014, 768380. [Google Scholar] [CrossRef]
- Raj, R.S.; Bonney, E.A.; Phillippe, M. Influenza, immune system, and pregnancy. Reprod. Sci. 2014, 21, 1434–1451. [Google Scholar] [CrossRef] [Green Version]
- Uchide, N.; Ohyama, K.; Bessho, T.; Takeichi, M.; Toyoda, H. Possible roles of proinflammatory and chemoattractive cytokines produced by human fetal membrane cells in the pathology of adverse pregnancy outcomes associated with influenza virus infection. Mediat. Inflamm. 2012, 2012, 270670. [Google Scholar] [CrossRef]
- Uchide, N.; Ohyama, K.; Bessho, T.; Yuan, B.; Yamakawa, T. Apoptosis in cultured human fetal membrane cells infected with influenza virus. Biol. Pharm. Bull. 2002, 25, 109–114. [Google Scholar] [CrossRef] [Green Version]
- Uchide, N.; Ohyama, K.; Yuan, B.; Sano, T.; Bessho, T.; Yamakawa, T. Differential mRNA expression of inflammatory cytokines in cultured human fetal membrane cells responding to influenza virus infection. Biol. Pharm. Bull. 2002, 25, 239–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zahedi-Amiri, A.; Sequiera, G.L.; Dhingra, S.; Coombs, K.M. Influenza a virus-triggered autophagy decreases the pluripotency of human-induced pluripotent stem cells. Cell Death Dis. 2019, 10, 337. [Google Scholar] [CrossRef] [PubMed]
- Jewett, J.F. Influenza pneumonia at term. N. Engl. J. Med. 1974, 291, 256–257. [Google Scholar] [PubMed] [Green Version]
- Lieberman, R.W.; Bagdasarian, N.; Thomas, D.; Van De Ven, C. Seasonal influenza A (H1N1) infection in early pregnancy and second trimester fetal demise. Emerg. Infect. Dis. 2011, 17, 107–109. [Google Scholar] [CrossRef] [PubMed]
- McGregor, J.A.; Burns, J.C.; Levin, M.J.; Burlington, B.; Meiklejohn, G. Transplacental passage of influenza A/Bangkok (H3N2) mimicking amniotic fluid infection syndrome. Am. J. Obs. Gynecol. 1984, 149, 856–859. [Google Scholar] [CrossRef]
- Yawn, D.H.; Pyeatte, J.C.; Joseph, J.M.; Eichler, S.L.; Garcia-Bunuel, R. Transplacental transfer of influenza virus. JAMA 1971, 216, 1022–1023. [Google Scholar] [CrossRef]
- Perdigao, A.C.; de Carvalho Araujo, F.M.; de Melo, M.E.; de Lemos, D.Q.; Cavalcanti, L.P.; Ramalho, I.L.; Araujo, L.C.; Sousa, D.M.; Siqueira, M.M.; Guedes, M.I. Post-pandemic influenza a (H1N1) 2009 virus infection in pregnant women in Ceara, Brazil. Influenza Other Respir. Viruses 2015, 9, 293–297. [Google Scholar] [CrossRef] [Green Version]
- Memoli, M.J.; Harvey, H.; Morens, D.M.; Taubenberger, J.K. Influenza in pregnancy. Influenza Other Respir. Viruses 2013, 7, 1033–1039. [Google Scholar] [CrossRef]
- Rasmussen, S.A.; Jamieson, D.J.; Bresee, J.S. Pandemic influenza and pregnant women. Emerg. Infect. Dis. 2008, 14, 95–100. [Google Scholar] [CrossRef]
- Cerbulo-Vazquez, A.; Figueroa-Damian, R.; Arriaga-Pizano, L.A.; Hernandez-Andrade, E.; Mancilla-Herrera, I.; Flores-Mejia, L.A.; Arteaga-Troncoso, G.; Lopez-Macias, C.; Isibasi, A.; Mancilla-Ramirez, J. Pregnant women infected with pandemic H1N1pdm2009 influenza virus displayed overproduction of peripheral blood CD69+ lymphocytes and increased levels of serum cytokines. PLoS ONE 2014, 9, e107900. [Google Scholar] [CrossRef] [Green Version]
- Pazos, M.; Sperling, R.S.; Moran, T.M.; Kraus, T.A. The influence of pregnancy on systemic immunity. Immunol. Res. 2012, 54, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Januar, V.; Desoye, G.; Novakovic, B.; Cvitic, S.; Saffery, R. Epigenetic regulation of human placental function and pregnancy outcome: Considerations for causal inference. Am. J. Obs. Gynecol. 2015, 213 (Suppl. 4), S182–S196. [Google Scholar] [CrossRef]
- Shalaby, F.; Rossant, J.; Yamaguchi, T.P.; Gertsenstein, M.; Wu, X.F.; Breitman, M.L.; Schuh, A.C. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 1995, 376, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Bolon, B. Pathology Analysis of the Placenta; Academic Press: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Wong, M.-T.; Chen, S.S. Hepatitis C Virus Subverts Human Choline Kinase-α To Bridge Phosphatidylinositol-4-Kinase IIIα (PI4KIIIα) and NS5A and Upregulates PI4KIIIα Activation, Thereby Promoting the Translocation of the Ternary Complex to the Endoplasmic Reticulum for Viral Replication. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Frabutt, D.A.; Wang, B.; Riaz, S.; Schwartz, R.C.; Zheng, Y.-H. Innate Sensing of Influenza A Virus Hemagglutinin Glycoproteins by the Host Endoplasmic Reticulum (ER) Stress Pathway Triggers a Potent Antiviral Response via ER-Associated Protein Degradation. J. Virol. 2017, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, J.; Nakatsu, F.; Baskin, J.M.; De Camilli, P. Plasticity of PI4KIIIα interactions at the plasma membrane. EMBO Rep. 2015, 16, 312–320. [Google Scholar] [CrossRef]
- De Marco, M.C.; Martin-Belmonte, F.; Kremer, L.; Albar, J.P.; Correas, I.; Vaerman, J.P.; Marazuela, M.; Byrne, J.A.; Alonso, M.A. MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells. J. Cell Biol. 2002, 159, 37–44. [Google Scholar] [CrossRef]
- Bernard, N.J.; O’Neill, L.A. Mal, more than a bridge to MyD88. Iubmb Life 2013, 65, 777–786. [Google Scholar] [CrossRef]
- Alonso, M.A.; Weissman, S.M. cDNA cloning and sequence of MAL, a hydrophobic protein associated with human T-cell differentiation. Proc. Natl. Acad. Sci. USA 1987, 84, 1997–2001. [Google Scholar] [CrossRef] [Green Version]
- Rancano, C.; Rubio, T.; Correas, I.; Alonso, M.A. Genomic structure and subcellular localization of MAL, a human T-cell-specific proteolipid protein. J. Biol. Chem. 1994, 269, 8159–8164. [Google Scholar]
- Millan, J.; Alonso, M.A. MAL, a novel integral membrane protein of human T lymphocytes, associates with glycosylphosphatidylinositol-anchored proteins and Src-like tyrosine kinases. Eur. J. Immunol. 1998, 28, 3675–3684. [Google Scholar] [CrossRef]
- Anton, O.; Batista, A.; Millan, J.; Andres-Delgado, L.; Puertollano, R.; Correas, I.; Alonso, M.A. An essential role for the MAL protein in targeting Lck to the plasma membrane of human T lymphocytes. J. Exp. Med. 2008, 205, 3201–3213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anton, O.M.; Andres-Delgado, L.; Reglero-Real, N.; Batista, A.; Alonso, M.A. MAL protein controls protein sorting at the supramolecular activation cluster of human T lymphocytes. J. Immunol. 2011, 186, 6345–6356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaud, G.; Lesourne, R.; Love, P.E. Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 2018, 18, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Albano, F.; Vecchio, E.; Renna, M.; Iaccino, E.; Mimmi, S.; Caiazza, C.; Arcucci, A.; Avagliano, A.; Pagliara, V.; Donato, G.; et al. Insights into Thymus Development and Viral Thymic Infections. Viruses 2019, 11, 836. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.J.; Geng, Y.; Cho, H.; Li, S.; Giri, P.K.; Felio, K.; Wang, C.R. Differential requirements for the Ets transcription factor Elf-1 in the development of NKT cells and NK cells. Blood 2011, 117, 1880–1887. [Google Scholar] [CrossRef] [Green Version]
- Lai, H.Y.; Hsu, L.W.; Tsai, H.H.; Lo, Y.C.; Yang, S.H.; Liu, P.Y.; Wang, J.M. CCAAT/enhancer-binding protein delta promotes intracellular lipid accumulation in M1 macrophages of vascular lesions. Cardiovasc. Res. 2017, 113, 1376–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szabo-Fresnais, N.; Blondeau, J.P.; Pomerance, M. Activation of the cAMP pathway synergistically increases IL-1-induced IL-6 gene expression in FRTL-5 thyroid cells: Involvement of AP-1 transcription factors. Mol. Cell Endocrinol. 2008, 284, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Mesnard-Rouiller, L.; Bismuth, J.; Wakkach, A.; Poea-Guyon, S.; Berrih-Aknin, S. Thymic myoid cells express high levels of muscle genes. J. Neuroimmunol. 2004, 148, 97–105. [Google Scholar] [CrossRef]
- Lee, Y.; Clinton, J.; Yao, C.; Chang, S.H. Interleukin-17D Promotes Pathogenicity During Infection by Suppressing CD8 T Cell Activity. Front. Immunol. 2019, 10, 1172. [Google Scholar] [CrossRef] [Green Version]
- Kushwah, R.; Oliver, J.R.; Wu, J.; Chang, Z.; Hu, J. Elf3 regulates allergic airway inflammation by controlling dendritic cell-driven T cell differentiation. J. Immunol. 2011, 187, 4639–4653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copie-Bergman, C.; Plonquet, A.; Alonso, M.A.; Boulland, M.L.; Marquet, J.; Divine, M.; Moller, P.; Leroy, K.; Gaulard, P. MAL expression in lymphoid cells: Further evidence for MAL as a distinct molecular marker of primary mediastinal large B-cell lymphomas. Mod. Pathol. 2002, 15, 1172–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreakos, E.; Sacre, S.M.; Smith, C.; Lundberg, A.; Kiriakidis, S.; Stonehouse, T.; Monaco, C.; Feldmann, M.; Foxwell, B.M. Distinct pathways of LPS-induced NF-kappa B activation and cytokine production in human myeloid and nonmyeloid cells defined by selective utilization of MyD88 and Mal/TIRAP. Blood 2004, 103, 2229–2237. [Google Scholar] [CrossRef] [PubMed]
- Puertollano, R.; Martin-Belmonte, F.; Millan, J.; de Marco, M.C.; Albar, J.P.; Kremer, L.; Alonso, M.A. The MAL proteolipid is necessary for normal apical transport and accurate sorting of the influenza virus hemagglutinin in Madin-Darby canine kidney cells. J. Cell Biol. 1999, 145, 141–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, S.; Zimman, A.; Gao, D.; Byzova, T.V.; Podrez, E.A. TLR2 Plays a Key Role in Platelet Hyperreactivity and Accelerated Thrombosis Associated With Hyperlipidemia. Circ. Res. 2017, 121, 951–962. [Google Scholar] [CrossRef]





| Target Gene | Accession Number | Primer Sequence |
|---|---|---|
| Reference genes | ||
| Gpi1 | NM_008155 | F: CGAACACGGCCAAAGTGAAA |
| R: AGCTGCTCGAAGTGGTCAAA | ||
| Hprt | NM_013556 | F: AGTCCCAGCGTCGTGATTAG |
| R: TCTCGAGCAAGTCTTTCAGTCC | ||
| Taldo1 | NM_011528 | F: TTGGCTGCACAACGAAGACC |
| R: ATTCGTTCCGTGAGCATCCG | ||
| Thymus Canonical Pathway Genes | ||
| Acta1 | NM_001272041 | F: CGCCAGCCTCTGAAACTAGA |
| Calcium (Ca2+) signaling | R: ACGATGGATGGGAACACAGC | |
| Cldn3 | NM_009902 | F: GCAAGGACTACGTCTGAGGG |
| Tight junction | R: ACTGTGTGTCGTCTGTCACC | |
| Mylpf | NM_016754 | F: ACCACGGTATGTTAAGGGCTG |
| Hepatic fibrosis | R: CCTTCTTGGGTGCCATGTCTTA | |
| Agt | NM_007428 | F: GTTGGCGCTGAAGGATACACA |
| Neuroprotective | R: GACCCAGGTCAAGATGCAGAA | |
| Thymus Immune Systems Genes | ||
| Mal | NM_001171187 | F: GTGAGTTTGATGCAGCCTACC |
| R: CCACTGCGGCGATGTTTTC | ||
| Mal2 | NM_178920 | F: GGACGTACTCCGGAGCTTTC |
| R: AGCTGTCACCGACACAAACA | ||
| Placenta Genes | ||
| Tmem150A | NM_144916.3 | F: TGAACAAGGGGGCCCTAAGA |
| R: AGATGAGGGCCACCATAACAG | ||
| Ppbp4 | NM_023785.3 | F: TGCTGATGTGGAAGTGATAGCC |
| R: GAAGCAGCTGGTCAGTAACCT | ||
| Ppbp8 | NM_023785.3 | F: ACAGCTGGAAAATCTGATGGCA |
| R: CTCCTGGCCTGTACACATTCA | ||
| A/California/07/2009(H1N1) | ||
| Matrix | NC_026431.1 | F: AGATGAGTCTTCTAACCGAGGTCG |
| R: TGCAAAGACACTTTCCAGTCTCTG | ||
| Sexing genes | ||
| Sry | NM_011564.1 | F: CTGGAGCTCTACAGTGATGA |
| R: CAGTTACCAATCAACACATCAC | ||
| Myog (control) | M95800.1 | F: TTACGTCCATCGTGGACAGCAT |
| R: TGGGCTGGGTGTTAGTCTTAT | ||
| Day of Inoculation | E3.5 | E7.5 | E12.5 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | Influenza | p | Control | Influenza | p | Control | Influenza | p | |
| Number of litters | 3 | 5 | 8 | 9 | 5 | 5 | |||
| Total number of fetuses | 26 | 43 | 59 | 59 | 36 | 44 | |||
| Average number of fetuses per litter ± standard error of the mean (SEM) | 8.7 ± 0.7 | 8.6 ± 0.5 | 0.9456 | 7.38 ± 0.75 | 6.56 ± 0.78 | 0.4768 | 7.2 ± 0.6 | 8.8 ± 0.8 | 0.1622 |
| Average fetal weight per litter (g) ± SEM | 1.16 ± 0.21 | 1.12 ± 0.07 | 0.8317 | 1.20 ± 0.02 | 1.03 ± 0.09 | 0.0674 | 1.25 ± 0.06 | 1.17 ± 0.04 | 0.2862 |
| Average fetal weight (g) ± SEM | 1.12 ± 0.06 | 1.14 ± 0.01 | 0.7844 | 1.21 ± 0.02 | 1.02 ± 0.03 | <0.0001 | 1.23 ± 0.02 | 1.16 ± 0.01 | 0.0067 |
| Average placental weight per litter (g) ± SEM | 0.13 ± 0.01 | 0.11 ± 0.002 | 0.0364 | 0.12 ± 0.01 | 0.10 ± 0.01 | 0.1432 | 0.11 ± 0.02 | 0.10 ± 0.01 | 0.4901 |
| Average placental weight (g) ± SEM | 0.12 ± 0.005 | 0.11 ± 0.002 | 0.0008 | 0.121 ± 0.01 | 0.101 ± 0.003 | 0.0013 | 0.105 ± 0.01 | 0.098 ± 0.003 | 0.2740 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Campen, H.; Bishop, J.V.; Abrahams, V.M.; Bielefeldt-Ohmann, H.; Mathiason, C.K.; Bouma, G.J.; Winger, Q.A.; Mayo, C.E.; Bowen, R.A.; Hansen, T.R. Maternal Influenza A Virus Infection Restricts Fetal and Placental Growth and Adversely Affects the Fetal Thymic Transcriptome. Viruses 2020, 12, 1003. https://doi.org/10.3390/v12091003
Van Campen H, Bishop JV, Abrahams VM, Bielefeldt-Ohmann H, Mathiason CK, Bouma GJ, Winger QA, Mayo CE, Bowen RA, Hansen TR. Maternal Influenza A Virus Infection Restricts Fetal and Placental Growth and Adversely Affects the Fetal Thymic Transcriptome. Viruses. 2020; 12(9):1003. https://doi.org/10.3390/v12091003
Chicago/Turabian StyleVan Campen, Hana, Jeanette V. Bishop, Vikki M. Abrahams, Helle Bielefeldt-Ohmann, Candace K. Mathiason, Gerrit J. Bouma, Quinton A. Winger, Christie E. Mayo, Richard A. Bowen, and Thomas R. Hansen. 2020. "Maternal Influenza A Virus Infection Restricts Fetal and Placental Growth and Adversely Affects the Fetal Thymic Transcriptome" Viruses 12, no. 9: 1003. https://doi.org/10.3390/v12091003