Abstract
Aims
In 2012, Italian kiwifruit orchards were hit by a serious root disease of unknown aetiology (kiwifruit decline, KD) that still causes extensive damage to the sector. While waterlogging was soon observed to be associated with its outbreak, the putative role of soil microbiota remains unknown. This work investigates the role of these two factors in the onset of the disease.
Methods
Historical rainfall data were analysed to identify changes that might explain KD outbreak and mimic the flooding conditions required to reproduce the disease in a controlled environment. A greenhouse experiment was thus designed, and vines were grown in either unsterilized (U) or sterilized (S) soil collected from KD-affected orchards, and subjected (F) or not (N) to artificial flooding. Treatments were compared in terms of mortality rate, growth, and tissue modifications.
Results
KD symptoms were only displayed by FU-treated vines, with an incidence of 90%. Ultrastructural observations detected tyloses and fibrils in the xylem vessels of all plants, irrespective of the treatment. Phytopythium vexans and Phytopythium chamaehyphon, isolated from roots of FU plants, emerged as the associated microorganisms.
Conclusions
We succeeded in reproducing KD under controlled conditions and confirmed its association with both waterlogging and soil-borne microorganism(s).
Similar content being viewed by others
Introduction
Kiwifruit orchards cover 0.4% of Italy’s agricultural land (around 26,400 ha), and the country is the second largest exporter, meeting over 15% of the global demand for this product (FAOSTAT 2017). Since 2012, a new threat named kiwifruit vine decline (KD) imperils Italian production (Tacconi et al. 2015). It appeared after 40 years of successful kiwifruit cultivation without any relevant phytosanitary problem, except for bacterial canker, caused by Pseudomonas syringae pv. actinidiae, affecting kiwifruit production since 2008. To date, kiwifruit vine decline (KD) has destroyed over 10% of all kiwifruit orchards (Veneto 1600 ha, Piedmont 1000 ha, Lazio 300 ha, Friuli Venezia Giulia 60 ha, and a few areas in Calabria; Sorrenti et al. 2019). However, unofficial sources estimated that more than 25% of the Italian kiwifruit growing areas have already been irreversibly compromised. To our knowledge, the disease is still limited to Italy but there are several reports of similar vine decline disorders or diseases in other countries, which can probably be ascribed to KD: Turkey (Akilli et al. 2011; Kurbetli and Ozan 2013; Polat et al. 2017), Japan (Huang and Qi 1998), China (Wang et al. 2015) and New Zealand (Reid et al. 1992).
The most obvious sign of KD is a rapid, sudden and irreversible wilting of the plant. No symptoms in the canopy are displayed until the summer heat waves occur (July – August in the Northern hemisphere), then the affected vines usually die within a few weeks. Symptoms throughout the canopy are associated with extensive damage to the root system. Coarse roots usually display a reddish discoloration under the cortex and their external cylinder easily detaches from the core, giving them a “rat-tail” appearance (Online resource 1). The greatest damage directly caused by KD is the almost complete decay of the white feeding roots. When feeding roots do not deteriorate, they usually remain localized within the top 5–10 cm of the soil (Tacconi et al. 2015). Infested land has never shown sign of recovery, forcing several farmers to shift to other crops (Tacconi et al. 2019).
KD is most frequently observed in silty soils with poor drainage, which are extremely susceptible to submersion (Sorrenti et al. 2019). Waterlogging is believed to be one of the main factors involved in the disease (Tacconi et al. 2015; Tosi et al. 2015; Sorrenti et al. 2019), although cases of KD have been reported in well-drained soils where flooding is rare, as well as in orchards where raised bed farming techniques were employed (Sorrenti et al. 2019).
Initially, most authors believed waterlogging to be the main cause of KD, and assumed that pathogens played a role of little or no importance, given that no direct link has been established between symptom appearance and presence of a given microorganism (Tacconi et al. 2014; Sorrenti et al. 2016; Fontana 2016). However, agronomic approaches focusing only on water management practices have failed to explain why Italian farmers are suddenly faced with such a large-scale epidemic after 40 years of kiwifruit cultivation (Sorrenti et al. 2019). The involvement of a biotic stress factor in KD was first proposed by Tacconi et al. (2015), but only recently confirmed by some studies (Donati et al. 2020; Spigaglia et al. 2020; Prencipe et al. 2020). However, rather than a unique pathogen, several microorganisms have been currently associated with the disease, among them: Fungi belonging to the Cylindrocarpon/Ilyonectria complex and Desarmillaria genera; Oomycetes of the Phytophthora, Pythium, and Phytopythium genera; and Bacteria of the Clostridium genus (Tosi et al. 2015; Tacconi et al. 2015; Donati et al. 2020; Spigaglia et al. 2020; Prencipe et al. 2020). Today, KD has been addressed either as a physiological response of kiwi plants to waterlogging (Tacconi et al. 2014; Tosi et al. 2015; Fontana 2016; Sorrenti et al. 2019), or as a disease caused by one or more soil-borne pathogens (Donati et al. 2020; Spigaglia et al. 2020; Prencipe et al. 2020), but a work specifically addressing the interaction of these two factors is still missing. This work aims at clarifying KD aetiology, investigating the role played by microorganisms and waterlogging in its onset under controlled conditions mimicking an open field environment.
Material and methods
A preliminary analysis of historical rainfall data was performed to define a flooding and drying pattern that could reliably mimic the waterlogging conditions occurring in KD-affected areas. For this investigation, both the duration and relative frequency of wet spells immediately before and after the first outbreak of KD (2012) were taken into account.
The experiments then carried out included: i) greenhouse trials aimed at discriminating between the effect of flooding and soil-borne pathogens on both symptom appearance and plant growth; ii) ultrastructural observations to find signs of cell alteration caused by the disease; iii) isolation of Fungi and Oomycetes to identify KD-associated microorganisms.
A further analysis of historical rainfall data was subsequently carried out to investigate possible correlations in three local case-studies between disease outbreak and changes in rainfall patterns over the last two decades. In particular, the research focused on changes in seasonal trends, rainfall event concentration and number of floods in both diseased and unaffected sites.
Greenhouse experiments
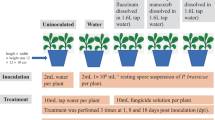
A greenhouse trial was set up to test the role of waterlogging and/or biotic factors in the aetiology of KD under controlled conditions. Three experimental conditions provided for different water stress conditions (flooding and non-flooding) with and without soil sterilization, as summarised in Fig. 1. The experimental conditions are defined as follows: NU are non-flooded (N) plants grown in unsterilized (U) soil; FU and FS are vines subjected to waterlogging (F), which were planted in non-sterile (U) and sterile (S) soil, respectively. FS- plants were considered as a negative control because preliminary investigations revealed that they displayed the same response as non-flooded vines growing in sterile soil in terms of mortality, canopy and root growth (details in Online resources 5 and 6).
Overview of the greenhouse trials and their timeline. The sequence of processes is depicted in grey; tick boxes indicate the processes carried out on the different experimental conditions. Grey circles (1,2,3) are the flooding stages. FS (green) are flooded (F) plants grown in sterile (S) soil; FU (orange) are flooded (F) vines planted in non-sterile soil (U); NU (blue) are non-flooded (N) plants in non-sterile (U) soil. In yellow, observation period for symptoms of wilting. In purple, sample collection for ultrastructural observations; in red, for biomass evaluation; in light blue, for the isolation of Fungi/Oomycetes
Soil collection and plant material
Soil samples were collected near symptomatic plants in 3 sites of the Friuli Venezia Giulia region (FVG, North-eastern Italy) (Table 1). Topsoil (5 to 10 cm) was discarded, keeping only the soil surrounding the roots displaying “rat-tail” symptoms. The samples were then mixed together in equal proportions and 1/3 of the resulting mixture was autoclaved at 120 °C for 20 min.
Eight-month-old micropropagated plantlets of A. deliciosa (A.Chev.) C.F.Liang & A.R.Ferguson cv. Hayward rooted in 250 ml-large peat blocks were used for the experiment. Twenty vines were used for each experimental condition. Just before transplanting, 1/4 of the rooted peat block was sliced horizontally to induce the emission of new feeding roots and allow the entrance of potential pathogens. Plants were then transplanted into black plastic pots, 6.5 l in volume. Three additional plants were potted into sterile peat and used as controls for the ultrastructural observations (C, ultrastructural control).
Water stress treatments
In the NU experimental condition, the soil moisture was kept slightly below field capacity, whereas both FU and FS plants were subjected to two flooding cycles. Each flooding cycle consisted of a number of flooding stages, made up of a flooding and a subsequent drainage step. Flooding was performed to induce symptom expression; the duration and frequency of this stage was estimated on the ground of the rainfall distribution recorded during the early vegetative season of kiwifruit vines in two KD-affected areas (Table 1, San Giovanni di Casarsa and San Giorgio della Richinvelda) between the first outbreak of the disease (2012) and the start of trial (2016), (see Section Historical rainfall data analysis, for details). The number of flooding stages in a flooding cycle was estimated based on the results obtained in the preliminary trials and set to 3, since most of plants died within this course of events (Online resource 6). A flooding cycle therefore had the following pattern: 3f-4d, 4f-4d and 5f-5d where #f is the number of consecutive days of flooding and #d is the duration of the following drainage step (Online resource 2). Flooding was mimicked by submerging the pots in buckets full of tap water with the water level maintained at least 1 cm above the soil surface. Only C plants were kept under optimal water condition for the entire duration of the trial.
Timeline
Following is a brief outline of the timeline for the operations, which is described in greater detail in Fig. 1. In late July 2017, kiwifruit plants were transplanted into sterilized or unsterilized soil collected one week beforehand. The vines were grown under optimal watering conditions until the beginning of the first flooding cycle (late September, 2017). Only FS and FU plants were flooded, while NU vines were watered regularly to keep the soil moisture just below field capacity, Online resource 2. After the first flooding cycle, all plants were kept at optimal water conditions until bud breaking. Three weeks after this stage, the vines were irrigated so as to replicate the rainfall events recorded daily in an unaffected location (Udine): when daily rainfall was less than or equal to 15 mm/d, irrigation was applied in a single event, otherwise the plants were watered twice (morning and evening, see Online resource 2 for further details). Each drip irrigation supplied water until the field capacity was exceeded by 200 ml. At the end of July 2018, a second flooding cycle was carried out following the same schedule as the first one.
Plants were constantly monitored over the whole trial period and the canopy was examined daily during the flooding cycles to detect any symptoms. Sampling for ultrastructural observation and isolation was performed as outlined in Fig. 1.
Sampling protocols and data analysis
Mortality rate and dry biomass measurements
Mortality rate was measured daily during both flooding cycles, considering as dead those plants whose canopy was completely wilted. Leaf and root biomass were measured to assess the effect of each experimental conditions and their interaction. Only roots that were directly in contact with the soil mixture, hence that had extended beyond the original peat block, were collected. These samples were then gently rinsed under running tap water to remove all soil residues. Roots and leaves were then oven-dried (60 °C for 4 days) before weighting. One-way ANOVA using Rstudio (Team 2013) was performed to compare the dry biomass and leaf/root ratio in the various experimental conditions.
Ultrastructural observations
Ultrastructural observations were carried out to detect cellular modifications induced by experimental conditions, and vascular pathogens or pathogen-specific changes in plant tissues. Fully-expanded leaves at the 6°-7° internode (60 cm height) were randomly harvested from three plants per each experimental condition. The three unstressed plants growing in sterile peat (C) were used as negative controls. Segments (3–4 mm in length) of leaf tissues including lateral veins and surrounding parenchyma cells were embedded in Epon-Araldite epoxy resin; ultrathin (60–70 nm) sections were cut with an ultramicrotome (Reichert Leica Ultracut E ultramicrotome, Leica Microsystems, Wetzlar, Germany) and collected on 200 mesh uncoated copper grids as described by Tarquini et al. (2019). Sections were then stained with UAR-EMS (uranyl acetate replacement stain) (Electron Microscopy Sciences, Fort Washington, PA, USA) and observed under a PHILIPS CM 10 (FEI, Eindhoven, The Netherlands) transmission electron microscope (TEM), operated at 80 kV, and equipped with a Megaview G3 CCD camera (EMSIS GmbH, Münster, Germany). Five non-serial cross-sections from each sample were analysed.
Fungi and Oomycetes isolation and identification
Although a role of bacteria in the aetiology of the disease could not be ruled out, we decided to focus on Fungi and Oomycetes because they were the organisms most likely involved, according to the epidemic patterns and the information available at the beginning of this project. Fungi and Oomycetes were isolated from the roots of plants displaying KD symptoms. Only roots with a diameter of less than 5 mm were used. The samples were rinsed under running tap water, surface sterilized in sodium hypochlorite 2% for 1 min and rinsed twice in sterilized water for 2 min. The overall sterilization process was repeated twice. Eight slices of root tissue were cut 3–10 mm above the rotting/healthy tissue interface of each plant. The slices were placed into two 90 mm Petri dishes containing potato dextrose agar (PDA; Oxoid Ltd) and streptomycin (100 μg/ml; Sigma-Aldrich). The Petri dishes thus inoculated were incubated in a growth chamber and kept in the dark for 48 h at 24 °C. Each fungal colony was sub-cultured at least five times.
After isolation, the colonies were grouped according to macroscopic features (colour, margin shape and growth rate) to determine morphological groups and successively subjected to molecular analysis. A maximum of 300 mg mycelia was collected by scraping the surface of 7- or 14-day-old colonies and stored at −20 °C until DNA extraction. High-molecular-weight DNA was extracted using the procedure described by Lecellier and Silar (1994) with slight modifications (Martini et al. 2009). The nuclear rDNA internal transcribed spacer regions (ITS1 and ITS2) and the 5.8S rRNA gene were amplified by PCR using the universal primers ITS1 and ITS4 (White et al. 1990). Amplifications were performed with an automated thermal cycler (MJ Research DNA Thermal Cycler PTC-100) in a 25-μL solution containing 200 μM each of the four dNTPs, 0.4 μM of each primer, 1.5 mM MgCl2, 0.625 units of DNA polymerase, GoTaq-Flexy (Promega), and 1 μL of 2 ng/μL diluted DNA. The PCR programme consisted of: 2 min at 94 °C followed by 35 cycles of 40 s at 94 °C; 30 s at 56 °C, 50 s at 72 °C and a final extension cycle of 7 min at 72 °C. The PCR reaction was checked by performing a 1% agarose gel electrophoresis in 1X TAE (60 min). Sequencing of ITS region and BLAST analysis were performed for fungal and oomycete species identification as detailed hereinafter. The PCR products of the isolates belonging to the less common morphological groups were directly Sanger sequenced; while the PCR products obtained from the most common groups were first analysed by PCR/RFLP method to confirm morphological grouping and possibly to demonstrate further genetic variability. ITS PCR products were digested using Tru1I and HinfI enzymes (Fermentas Ltd). Restriction enzymes were selected with the software CisSERS (Sharpe et al. 2016) using as inputs the ITS sequences of 23 different Fungi and Oomycetes genera gained from isolation attempts carried out in a preliminary work. Enzymes differing for the CG content of their restriction sites and generating in silico unique profiles for each genus of Fungi and Oomycetes were chosen for the actual RFLP analysis. RFLP patterns were visualized on 2% agarose gel electrophoresis in 1X TBE, and compared within each morphological group to differentiate the isolates into molecular groups. Within each molecular group, on average 40% of the isolates were selected for sequencing the ITS region on the basis of the plant of origin. The ITS1/ITS4 PCR products (about 500 to 600 bp for Fungi, 800-1000 bp for Oomycetes) were purified using a Wizard SV Gel and the PCR Clean-Up System Kit (Promega, Madison, WI, USA). Sanger sequencing was performed using the ITS4 primer by Genechron Laboratories, (Genechron S.r.l, Italy). Sequences were then compared with BLAST algorithms against NCBI GenBank database for taxonomical identification (Altschul et al. 1990).
Historical rainfall data analysis
Rainfall data from 1998 to 2018 recorded by the meteorological stations in Vivaro, Palazzolo and Udine (Friuli Venezia Giulia, FVG, North-eastern Italy), were downloaded from the OMNIA regional weather database (Cicogna et al. 2015) (Table 1). The first orchards to be affected by KD were located near Vivaro and Palazzolo (2012–2014), whereas to date, Udine still seems to be free of the disease. The first case of KD was officially diagnosed in FVG in 2014 (Savian et al. 2017), but by combining farmers’ accounts with Google Earth historical satellite data (Google LLC 2019) we were able to trace the first outbreak back to 2012 (Saro S., ERSA – Phytosanitary Service of Friuli Venezia Giulia region, personal communication).
In order to verify a possible link between KD and waterlogging, we examined rainfall data during the early vegetative (March–May) and ripening (August–October) stages of kiwifruit vines, given that during these two periods floods are usually more frequent and root decay is potentially more harmful considering the kiwifruit rooting patterns (Buwalda and Hutton 1988).
We investigated the pattern of rainfall events taking into account the duration of wet spells, the average time span between two consecutive wet periods (ET, used as a proxy of rainfall concentration over time), cumulative rainfall evolution (CR) and the frequency of flood-causing downpours (FD).
Firstly, the analysis of rainfall data in affected areas from the first outbreak of the disease (2012) to the beginning of the experiments (2016) allowed us to define the duration of the flooding and drainage steps used in the greenhouse trials. The number of wet spells (classified by length: 1, 2, ≥3 days) occurring during the early vegetative stage determined the reference duration of the flooding step, while mean ET determined that of the drainage step.
Secondly, the rainfall time series over the last two decades was analysed to identify changes in trends, rain intensity and distribution that could explain the sudden and widespread diffusion of the disease after 2012, considering data from both diseased and unaffected locations. A preliminary flat step analysis and piecewise regression was carried out on the cumulative rainfall (CR) time series to detect any change points (Mariani 2006; Chiaudani et al. 2008). The Theil-Sen nonparametric method was applied to identify any linear trends in CR data (Helsel 2005) and the Mann-Kendall test was subsequently used to assess the significance of the resulting slope (Mann 1945; Kendall 1975). Successively, frequencies of FD and mean ET before and after the first outbreak of the disease (2012) were compared in every location using the Mann-Whitney-Wilcoxon test.
The estimation of frequency of FD and mean ET was based on the following assumptions: i) a day was considered rainy when more than 1 mm fell, and ii) a downpour causes water excess when the rain fallen exceeds the flooding threshold. The latter was estimated as the amount of water needed to exceed the soil field capacity considering that in kiwifruit orchards the soil moisture is usually kept above the 65% of the available water content (Xiloyannis et al. 1997). Root depth was set at 30 cm from the soil surface, because this is the region where most of the superficial root system of kiwifruit resides. Available water content was estimated from soil texture composition using the SPAW models (Saxton and Willey 2005). Soil properties (texture, maximum water content and saturation volume) are listed in Table 1. All the analyses were performed using R statistical software (Team 2013).
Results
Definition of flooding cycles
Analyses of the duration of rainy spells during the period 2012 to 2016 revealed that most rainfall events in the affected areas (Palazzolo, Vivaro) lasted 1–2 days, although more prolonged rainy periods (≥ 3 consecutive days) were not rare and occurred at least twice a year (Fig. 2a, b). The time elapsing between two consecutive events averaged 4.8 days (Fig. 2c, d). In the light of these data, flooding conditions for the greenhouse experiments were set with a reference combination of 3 days flooding followed by 4 days of drainage. This flooding/drainage combination was used in the first flooding stage of the cycle; in the following ones, the duration of the flooding step was progressively increased, so that each flooding cycle consisted of three stages, which can be summarized as follows: 3f-4d; 4f-4d; 5f-5d.
Rainfall distribution parameters taken into account to define the flooding cycle. Rainfall data during the early vegetative stage (March–June) of kiwifruit vines growing in diseased areas (Palazzolo, a, c; Vivaro, b, d) from the first outbreak of KD (2012) to the beginning of the greenhouse trials (2016). The number of wet spells (a, b) is depicted in blue: the darkness of the shade is proportional to the duration of the rainy spell (1, 2, 3 days or more). In green, the average elapsed time (ET) between two consecutive rainfalls (c, d)
Symptoms, mortality rates, and biomass measurements
The experimental design proved successful in reproducing KD symptoms in a controlled environment. No signs of the disease were observed during the first flooding cycle in September 2017. Symptoms were first detected in July 2018 during the second flooding cycle, just after the first flooding stage (Fig. 3). Wilting always started from the basal leaves and quickly moved upwards, usually ending with the all leaves falling. Complete wilting occurred 7 to 10 days after the first symptoms appeared. The severity of wilting and the number of damaged plants increased as the flooding cycle progressed. No sign of recovery was ever observed once wilting began.
Evolution of wilting symptoms during the second flooding cycle (July 2018). a Watering regime (WR): blue triangles depict when NU (non-flooded, in non-sterile soil) vines were watered; light blue rectangles indicate for how many days the plants grown in sterile (FS) and non-sterile soil (FU) were waterlogged. b Mean (red line), maximum and minimum (light red shaded area) air temperature values inside the greenhouse. c-e Number of healthy (green) and wilted (orange) plants per experimental condition. f-h Images of representative plants for each experimental conditionat the end of the trial period
FS vines successfully colonized the entire pot, displaying healthy white feeding roots, whereas the roots of NU plants extended just a few centimetres beyond the peat block and, in a small number of cases, exhibited a partial rotting of the smaller feeding roots (Fig. 3f, g). On the other hand, FU plants displayed the classical KD symptoms: a serious deterioration of the feeding roots, rotting tissues in the smaller (<5 mm) coarse roots, a reddish discoloration beneath the cortex of coarse roots, and a rat-tail appearance (Fig. 3h).
By the end of the trial period, most (90%) FU plants revealed KD symptoms whereas FS and NU vines had none (Fig. 3). However, highly significant differences (P < 0.001) were observed between both root dry weight and leaf dry weight of FS and NU plants, as well as in their ratio (Fig. 4); the dry weight of FU vines was not measured, given their severe reduction in secondary roots and abundant loss of leaves. FS plants had approximately double the amount of roots and leaves respect to NU-treated ones, and also had a more balanced leaf/root ratio, almost half than that of NU vines.
Ultrastructural observations
Ultrastructural observations revealed tyloses and fibrils in the xylem vessels (Fig. 5) of all the treated vines (FS, NU and FU), but not the controls (C, non-flooded plants growing in sterile peat). Gums adhering to the cell walls were found in all samples. Tyloses and fibrils appeared to be more abundant in FU vines than in NU and FS ones. No pathogen-related structures (hyphae, conidia) were found in the xylem and likewise, no pathogen-related symptoms (such as xylem cell wall deterioration) were observed. Finally, no unusual alteration of the parenchyma and phloem cells was detected.
TEM micrographs of xylem vessels of A. deliciosa cv. Hayward subjected to different combinations of water supply and soil sterilization conditions. Whole cells (a-d) and details (e-f) of xylem vessels (Xy). Sections of plants grown in non-sterile (FU; a, e) and sterile soil (FS; c, g) and subjected to flooding; not subjected to flooding, and growing in non-sterile soil (NU; b, f) and sterile peat (c; d, h). Tyloses (t) are visible in (a, e and b), fibrils (fi) in (a, c and e). Gum deposits (arrows) are visible in all samples (a-h). Scale bar = 2 μm
Isolation results
One hundred twenty-eight isolates were obtained from the roots of the 18 symptomatic FU-treated plants. A total of 22 morphological groups were identified on the ground of their macroscopic characteristics. Successively, RFLP analysis on the ITS region allowed to easily differentiate isolates belonging to distinct morphological groups. Moreover, it was possible to further characterize the major morphological groups on the basis of their genetic variability (Online resources 3 and 4); for example, within the morphological group 1, 4 molecular groups (1A-D) were identified. After morphological and molecular grouping, 76 isolates were selected for Sanger sequencing of their ITS region. Sequence analysis of ITS region did not evidence relevant genetic variability at the species level within molecular groups and revealed 42 unique sequences ascribable to 23 different species. Families with the highest occurrence were Hypocreaceae, Ceratobasidiaceae, Pythiaceae, and Nectriaceae (56, 67, 89 and 100%, respectively), but as regards the species, only Fusarium solani (78%), Phytopythium chamaehyphon (61%) and Phytopythium vexans (56%) were frequently found, Fig. 6. F. solani presented several sequence types (8 unique sequences) but none of them had a high frequency of occurrence (less than 22%). As regards P. vexans, only one out of five sequence was slightly different (two SNPs) from all the others. All the sequences of P. chamaehyphon were identical. Information regarding isolation, including accession numbers of the isolates from this study, is reported in Online resource 8.
Historical rainfall data analysis
Descriptive statistics of rainfall time series revealed that the average cumulative rainfall (CR) during the early vegetative stage of kiwifruit vines differed quite extensively between the two diseased areas over the years 1998 to 2018: 281 mm in Palazzolo and 401 mm in Vivaro. As regards the ripening stage, the CR in the two areas was of 342 and 456 mm, respectively. The pluviometric regime in the unaffected area (Udine) had intermediated values, with an average of 350 mm during the early vegetative stage and 437 mm in the fruit ripening one. In 2013, rainfall peaks during the early vegetative stage were noteworthy both in terms of cumulative rainfall (1012 mm in Palazzolo, 1052 mm in Vivaro and 1129 mm in Udine Fig. 7a-c) and wet spell duration (Fig. 7).
Statistical analysis of historical rainfall data over the last two decades (1998–2018) in Palazzolo, Vivaro and Udine (North-eastern Italy). In green, data relative to the early vegetative stage (March–May); in orange, relative to ripening (August–October). a-c Cumulative rainfall (CR) and corresponding Theil-Sen trends (continuous lines) and observed data (dots and squares); the significance of the trend slopes is also shown (P < 0.05). d-f, j-l Number of flooding events (FD). g-i, m-o elapsed time (ET) between two consecutive rainfalls; from light to dark, the 10th, 50th and 90th percentile. The light blue background highlights the time interval since the first appearance of kiwifruit decline in FVG
Change point analysis identified no significant transition points in the CR data series. Overall, slight negative trends were detected in the CR of both early vegetative and ripening stages (Fig. 7a-c), which were generally not significant, except in two cases (Palazzolo during the vegetative stage, Udine during ripening).
Frequency of flood-causing downpours (FD) in the two affected locations differed extensively (an average of 4.76 and 6.24 events in Palazzolo during the early vegetative and ripening stages, respectively, vs. 7.33 and 8.67 in Vivaro) but was very similar between Vivaro and the unaffected site (i.e. Udine, with 7.57 and 10.24 events, respectively, Fig. 7d-f and Fig. 7j-l).
As regards inter-event time, the mean elapsed time (ET) between two consecutive wet spells was similar for all the locations: 4.5 and 4.9 days in Vivaro during the early vegetative and ripening stages, respectively, 4.9 and 5.1 days in Udine, 5.4 and 5.6 days in Palazzolo (Fig. 7g-k and Fig. 7m-o).
No increase in flood-causing downpours was observed nor in the frequency of rainy events; no significant differences in mean FD (Fig. 7d-f and Fig. 7j-l) and mean ET (Fig. 7g-k and Fig. 7m-o) were detected before and after first outbreak of KD in 2012 (Mann-Whitney, P > 0.15, Online resource 7).
Discussion
The aetiology of KD has been puzzling the Italian kiwifruit sector for the last seven years (Sorrenti et al. 2019). The swiftness with which this syndrome has spread across Italy and the astonishingly rapid progress that it has made throughout the affected areas would suggest the involvement of an aggressive alien pathogen; however the attempts to find the infective agent have failed so far.
The renowned susceptibility of kiwifruit vine to waterlogging (Savé and Serrano 1986; Smith et al. 1990; Reid and Petrie 1991; Reid et al. 1991, 1992), combined with the frequent occurrence of KD in poorly drained soils (Tacconi et al. 2014, 2015; Tosi et al. 2015; Sorrenti et al. 2016), misled the first researchers investigating its aetiology, who identified waterlogging as the main cause of KD. In particular, i) Smith et al. (1990) observed that flooding caused the cortex to separate from the central stele of the root, in a manner similar to that caused by KD; ii) a sudden plant wilting was described by Reid et al. (1991) right after floods caused by a cyclone; and iii) a reduction in root growth due to prolonged or transient flooding was reported by Savé and Serrano (1986) and Smith et al. (1990), respectively. Moreover, waterlogging changes the above/below ground biomass ratio, because shoots are only marginally affected whereas roots are strongly inhibited (Savé and Serrano 1986).
The hypothesis of waterlogging as the causal agent of KD however, did not hold up against the analysis of rainfall data series, which, as reported previously, failed to highlight any change over the last two decades that could explain the outbreak of KD. Nonetheless, it is interesting to note that even if the first KD outbreak in FVG dates back to 2012, the disease escalated in 2014 (Savian et al. 2017) during a hot and dry vegetative season preceded by the rainiest year in the last two decades (2013). Although the historical rainfall analysis approach used in this work did not employ soil water models to predict water stagnation, the indications were clear enough to discard the hypothesis of a major change in rainfall distribution as the sole cause of the first outbreaks of KD.
For the first time, our results clearly reveal that waterlogging is a prerequisite for the deteriorations of feeding roots and the onset of the decline syndrome, but also that a soil-borne pathogen must be present to induce the disease.
The greenhouse experiments were successful in reproducing unequivocal symptoms of KD in both the roots and canopy of A. deliciosa cv. Hayward. These results were consistent with those of the trials performed in the field, both in terms of kiwifruit vine physiology and environmental conditions.
The vines were planted in August, so as to take full advantage of the most active period for rooting (Buwalda and Hutton 1988). Moreover, the flooding cycles mimicked the rainfall conditions that usually occur in the Friuli-Venezia Giulia region. Moreover, three or more days of flooding have been reported as sufficient to weaken kiwifruit plants (Savé and Serrano 1986; Smith et al. 1990). Our ultrastructural observation of the lumina of the xylem vessels revealing the presence of tyloses and fibrils, confirmed the stressing effect of the flooding conditions employed during the trial. This is the first time that such structures have been reported in kiwi plants affected by KD, suggesting that xylem vessel occlusion may be a plant response to either pathogen infection or flooding stress. Tyloses and fibrils are indeed unspecific plant stress responses (De Micco et al. 2016) that have been linked to both flooding condition (Davison and Tay 1987) and/or pathogen infection (Dimond 1955; Davison 2014; De Micco et al. 2016).
By prolonging the flooding step as the trial progressed, we promoted the progressive expression of KD symptoms, allowing a better characterization of the disease. During the second flooding cycle, flooded plants grown in unsterilized soil (FU) suddenly die as observed in the field (Tosi et al. 2015; Tacconi et al. 2015), and their death is associated with a greatly unbalanced leaf/root ratio and high environmental temperatures. The results of this investigation confirm those obtained during the preliminary trial where, once again, 88% of the plants died when flooding was applied on unsterile soil (Online resources 5 and 6), thus strengthening the reliability of our protocol in reproducing KD.
Our results are consistent with a model where pathogen aggressiveness is enhanced by waterlogging. On one hand, our measurements revealed highly significant differences in the dry weight of roots and leaves (and in their ratio) harvested from plants growing in sterilized and non-sterile, indicating that the severe damage to the kiwifruit root system was caused by microorganism(s) present in soil. On the other, the evidence of obvious KD symptoms in FU vines and their absence in NU ones highlighted the dramatic effect of flooding and its ability to promote an unbalanced above/below ground biomass ration in the presence of pathogen infection. The reason for the marginal effect of waterlogging observed in plants grown in sterile soil can be ascribed to the flooding conditions employed, which were intentionally calibrated to induce bearable stress. Indeed, kiwifruit vines respond to root loss caused by transient waterlogging, i.e. lasting less than 5 days, by sprouting new roots (Smith et al. 1990). The hypothesis of a synergy between soil-borne pathogens and waterlogging is also supported by the larger amount of tyloses and fibrils found in the vessels of FU plants, as compared to the other experimental conditions and the controls.
The results of the isolations provided a first idea of the soil-borne microorganisms potentially associated with KD. Given the frequency of their occurrence, the major candidates would appear to be P. chamaehyphon, P. vexans and F. solani.
In the literature F. solani was shown to cause crown rot on kiwifruit only when forcefully introduced in the collar by wounding, while it seemed unable to cause fine root decay if inoculated by submerging kiwi roots under a suspension of hyphae and conidia (Porta-Puglia and Corazza 1985). However, it was reported as secondary invader; unable to cause root rot by itself, but able to accelerate the fine root decay when associated with Oomycetes (Huang and Qi 1998).
P. vexans was recently found pathogenic causing KD in Prencipe et al. (2020), and it was also described as pathogenic in Turkey in a disease similar to KD causing root and collar rot (Polat et al. 2017). Very recently P. chamaehyphon has been reported in Italy on kiwifruit affected by KD (Donati et al. 2020). Nevertheless, a closely related species, Phytopythium helicoides, has been previously associated with root and collar rot of kiwifruit in China (Wang et al. 2015).
The newly published studies underline the involvement of several pathogens as associated with KD (Donati et al. 2020; Spigaglia et al. 2020; Prencipe et al. 2020). Beside the above mentioned species of Phytopythium, also Phytophthora infestans, Phytophthora megasperma, and Desarmillaria tabescens were demonstrated to be pathogenic (Donati et al. 2020), together with two Clostridium species: C. bifermentans and C. subterminale (Spigaglia et al. 2020). The presence of multiple pathogens well relate with our isolation results, since we found more than one microorganism with high occurrence.
Considering our results and those from other recent studies on KD, Oomycetes seem to be a key taxa for the onset of KD not only because they were constantly isolated from diseased plants (Donati et al. 2020; Prencipe et al. 2020), but also because their life cycle (Toussoun et al. 1970) is in perfect agreement with the spreading pattern of the disease. This hypothesis finds further proof on the well-known root rot activity of Oomycetes on kiwifruit vines (Latham and Dozier 1989; Conn et al. 1991; Latorre et al. 1991; Stewart and McCarrison 1992; Akilli et al. 2011; Kurbetli and Ozan 2013; Çiftçi et al. 2016). However, we cannot exclude that also other microorganisms such as Bacteria (Spigaglia et al. 2020) and Fungi (Donati et al. 2020) might take advantage of the weakened status of the plant caused by waterlogging.
The results obtained allow us to speculate about the dynamics of the disease and the sudden appearance of the symptoms. The ability of kiwifruit vines to take up great amounts of water when available (Xiloyannis et al. 1993), together with the large size of its xylem vessels and the many pits, allows the plants to rapidly take up and relocate water where needed (Dichio et al. 1999, 2013). Kiwifruit vines can therefore endure consistent reductions in the size of their root system (up to 80%) before limiting their shoot growth rates (Black et al. 2011), as long as water is not a limiting factor. However, this can be a double-edged sword, since: i) the large vessels make kiwifruit more susceptible to vessel occlusion by tyloses and gums (De Micco et al. 2016), and ii) the great ability to take up and translocate water might mask a compromised root system. Therefore, kiwifruit vines could have developed an abundant canopy in springtime even in the presence of severe root deterioration. When the first heat waves occur, transpiration rate could then easily exceed uptake capability of the root system, leading to the sudden wilting of the whole plant.
In conclusion, this is the first comprehensive approach to KD and the first demonstration of the synergic, necessary interaction between waterlogging and soil-borne pathogens for the onset of this complex disease. We have also defined a reliable protocol to reproduce KD in a controlled environment.
Several aspects require further investigations to clarify the dynamics of this new disease. A better understanding of the root deterioration processes is needed to improve sampling times and strategy in both controlled-environment trials and open-field systems. Detection methods need to be defined, in order to promptly evaluate the health status of the vines and predict potential outbreaks of this disease. Analysis techniques that can measure water stress-related parameters may prove to be helpful, such as: i) remote and proximal sensing data analysis in near and far infrared spectra (Mahlein 2015), ii) measurement of stomatal conductance and leaf water potential (Schaffer and Andersen 1994), iii) measurements of ABA content in the leaves or fruits (Schaffer and Andersen 1994), or iv) measurements of photosynthesis performance/stress (Ashraf 2012).
Finally, a further characterization of the role played by each factor involved in root decay as well as pathogenicity tests to confirm Koch’s postulates are necessary to fully clarify the aetiology and sustain our preliminary indications about the involvement of one or more oomycete species in KD.
In this regard, metabarcoding analysis might prove to be a useful tool to investigate the microbiota associated with healthy, asymptomatic and diseased plants, with the aim to give us a deeper insight on the putative pathogens responsible for a complex disease as KD. Nowadays, all the putative pathogens are regarded as the “pathobiome”, an emerging concept which is receiving greater attention (Lamichhane and Venturi 2015; Abdelfattah et al. 2018) and that well describes the plurality of microorganisms associated with KD.
Indeed, rather than one disease caused by a single pathogen, which can be proved using Koch’s postulates, complex diseases involve the sequential, combined and cumulative effects of biotic and abiotic agents (Denman et al. 2018). In the case of KD, symptoms seem to be the result of a complex interaction between waterlogging, plant and soil-borne pathogens. For KD as for all the other complex diseases the fulfilment of all the Koch’s postulates is a difficult task, which can be achieved using a multi-faced methodological and conceptual approach (Wallace 1978; Lamichhane and Venturi 2015; Denman et al. 2018).
Abbreviations
- C:
-
experimental condition acronym for plants grown in sterilized peat and under optimal water condition
- CR:
-
cumulative rainfall (mm)
- ET:
-
elapsing time between two consecutive rainfall
- FD:
-
downpours causing flooding
- FS:
-
experimental condition acronym for flooded plants grown in sterilized soil
- FU:
-
experimental condition acronym for flooded plants grown in unsterilized soil
- KD:
-
Kiwifruit decline
- NU:
-
experimental condition acronym for non-flooded plants grown in unsterilized soil
References
Abdelfattah A, Malacrinò A, Wisniewski M, Cacciola SO, Schena L (2018) Metabarcoding: a powerful tool to investigate microbial communities and shape future plant protection strategies. Biol Control 120:1–10
Akilli S, Serçe ÇU, Zekaİ Katircioğlu Y, Karakaya A, Maden S (2011) Involvement of Phytophthora citrophthora in kiwifruit decline in Turkey. J Phytopathol 159:579–581
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Ashraf MA (2012) Waterlogging stress in plants: a review. African journal of agricultural reseearch 7:1976–1981
Black MZ, Patterson KJ, Minchin PEH, Gould KS, Clearwater MJ (2011) Hydraulic responses of whole vines and individual roots of kiwifruit (Actinidia chinensis) following root severance. Tree Physiol 31:508–518
Buwalda JG, Hutton RC (1988) Seasonal changes in root growth of kiwifruit. Sci Hortic 36:251–260
Chiaudani A, Berti A, Borin M, Mariani L (2008) Piecewise and strucchange: test of two methods of change point analysis in agroclimatology. In: S. Michele all’Adige, Italy: Rivista Italiana di Agrometeorologia
Cicogna A, Medeossi R, Bellan A, Gimona A, Stefanuto L, Micheletti S (2015) Omnia: a new tool for the management of meteorological data. Italian Journal of Agrometeorology 20:73–80
Çiftçi O, Serçe ÇU, Türkölmez Ş, Derviş S (2016) First report of Phytophthora palmivora causing crown and root rot of kiwifruit (Actinidia deliciosa) in Turkey. Plant Dis 100:210
Conn KE, Gubler WD, Mircetich SM, Hasey JK (1991) Pathogenicity and relative virulence of nine Phytophthora spp. from kiwifruit. Phytopathology 81:974–979
Davison EM (2014) Resolving confusions about jarrah dieback - don’t forget the plants. Australas Plant Pathol 43:691–701
Davison EM, Tay FCS (1987) The effect of waterlogging on infection of Eucalyptus marginata seedlings by Phytophthora cinnamomi. New Phytol 105:585–594
De Micco V, Balzano A, Wheeler EA, Baas P (2016) Tyloses and gums: a review of structure, function and occurrence of vessel occlusions. IAWA J 37:186–205
Denman S, Doonan J, Ransom-Jones E, Broberg M, Plummer S, Kirk S, Scarlett K, Griffiths AR, Kaczmarek M, Forster J, Peace A, Golyshin PN, Hassard F, Brown N, Kenny JG, McDonald JE (2018) Microbiome and infectivity studies reveal complex polyspecies tree disease in acute oak decline. The ISME Journal 12:386–399
Dichio B, Baldassarre R, Nuzzo V, Biasi R, Xiloyannis C (1999) Hydraulic conductivity and xylem structure in young kiwifruit vines. Acta Hortic:159–164
Dichio B, Montanaro G, Sofo A, Xiloyannis C (2013) Stem and whole-plant hydraulics in olive (Olea europaea) and kiwifruit (Actinidia deliciosa). Trees 27:183–191
Dimond AE (1955) Pathogenesis in the wilt diseases. Annu Rev Plant Physiol 6:329–350
Donati I, Cellini A, Sangiorgio D, Caldera E, Sorrenti G, Spinelli F (2020) Pathogens associated to kiwifruit vine decline in Italy. Agriculture 10:119
FAOSTAT (2017) Food and agriculture organization of the United Nations. Rome, Italy
Fontana E (2016) La moria del kiwi avanza verso il nord-ovest. www.freshplaza.it/article/4086587/la-moria-del-kiwi-avanza-verso-il-nord-ovest/
Google LLC (2019) Google earth pro. Google LLC
Helsel DR (2005) Nondetects and data analysis: statistics for censored environmental data. Wiley-Interscience, Hoboken, N.J
Huang Y, Qi P (1998) Studies on the cause of root rot of kiwifruit in Guangdong Province. Journal of South China Agricultural University 19:19–22
Kendall MG (1975) Rank correlation methods. 4th Edition, Charles Griffin, London
Kurbetli İ, Ozan S (2013) Occurrence of Phytophthora root and stem rot of kiwifruit in Turkey. J Phytopathol 161:887–889
Lamichhane JR, Venturi V (2015) Synergisms between microbial pathogens in plant disease complexes: a growing trend. Front Plant Sci 06
Latham AJ, Dozier WA (1989) Root rot of kiwi (Actinidia chinensis) caused by Pythium ultimum. Plant Dis 73:938
Latorre BA, Alvarez C, Ribeiro OK (1991) Phytophthora root rot of kiwifruit in Chile. Plant Dis 75:949–952
Lecellier G, Silar P (1994) Rapid methods for nucleic acids extraction from Petri dish-grown mycelia. Curr Genet 25:122–123
Mahlein A-K (2015) Plant disease detection by imaging sensors – parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis 100:241–251
Mann HB (1945) Nonparametric tests against trend. Econometrica 13:245–259. https://doi.org/10.2307/1907187
Mariani L (2006) Some methods for time series analysis in agrometeorology. Rivista Italiana di Agrometeorologia:48–55
Martini M, Musetti R, Grisan S, Polizzotto R, Borselli S, Pavan F, Osler R (2009) DNA-dependent detection of the grapevine fungal endophytes Aureobasidium pullulans and Epicoccum nigrum. Plant Dis 93:993–998
Polat Z, Awan QN, Hussain M, Akgül DS (2017) First report of Phytopytium vexans causing root and collar rot of kiwifruit in Turkey. Plant Dis 101:1058
Porta-Puglia A, Corazza L (1985) Fusarium solani (Mart.) Sacc. associato ad un marciume del colletto dell’Actinidia. Rivista di ortoflorofrutticoltura italiana 69:81–83
Prencipe S, Savian F, Nari L, Ermacora P, Spadaro D, Martini M (2020) First report of Phytopythium vexans causing decline syndrome of Actinidia deliciosa ‘Hayward’ in Italy. Plant Disease: PDIS-10-19-2101
Reid JB, Petrie RA (1991) Effects of soil aeration on root demography in kiwifruit. N Z J Crop Hortic Sci 19:423–432
Reid JB, Tate KG, Brown NS, Cheah LH (1991) Effects of flooding and alluvium deposition on kiwifruit (Actinidia deliciosa): 1. Early vine decline. New Zealand Journal of Crop and Horticultural Science 19:247–257
Reid JB, Tate KG, Brown NS (1992) Effects of flooding and alluvium deposition on kiwifruit (Actinidia deliciosa). N Z J Crop Hortic Sci 20:283–288
Savé R, Serrano L (1986) Some physiological and growth responses of kiwi fruit (Actinidia chinensis) to flooding. Physiol Plant 66:75–78
Savian F, Martini M, Borselli S, Saro S, Musetti R, Loi N, Firrao G (2017) Studies on kiwifruit decline, an emerging issue even for Friuli Venezia Giulia (Eastern Italy). J Plant Pathol 99:18
Saxton KE, Willey PH (2005) The SPAW model for agricultural field and pond hydrologic simulation. Watershed models: 400–435
Schaffer B, Andersen PC (eds) (1994) Handbook of environmental physiology of fruit crops. CRC Press, Boca Raton
Sharpe RM, Koepke T, Harper A, Grimes J, Galli M, Satoh-Cruz M, Kalyanaraman A, Evans K, Kramer D, Dhingra A (2016) CisSERS: customizable in silico sequence evaluation for restriction sites (M Prasad, Ed.). PLOS ONE 11:e0152404
Smith GS, Judd MJ, miller SA, Buwalda JG (1990) Recovery of kiwifruit vines from transient waterlogging of the root system. New Phytol 115:325–333
Sorrenti G, Toselli M, Reggidori G, Spinelli F, Tosi L, Giacopini A, Tacconi G (2016) Implicazioni della gestione idrica nella “moria del kiwi” del veronese. Frutticoltura 3:1–7
Sorrenti G, Tacconi G, Tosi L, Vittone G, Nari L, Savian F, Saro S, Ermacora P, Graziani S, Toselli M (2019) Avanza la “moria del kiwi”: evoluzione e primi riscontri della ricerca. Frutticoltura 2:34–42
Spigaglia P, Barbanti F, Marocchi F, Mastroleo M, Baretta M, Ferrante P, Caboni E, Lucioli S, Scortichini M (2020) Clostridium bifermentans and C. subterminale are associated with kiwifruit vine decline, known as moria, in Italy. Plant Pathol 69:765–774
Stewart A, McCarrison AM (1992) Pathogenicity and relative virulence of seven Phytophthora species on kiwifruit. N Z J Crop Hortic Sci 19:73–76
Tacconi G, Giacopini A, Tosi L (2014) La Moria Del Kiwi Nel Veronese. Kiwi informa Aprile/Giugno:5–23
Tacconi G, Paltrinieri S, Mejia JF, Fuentealba SP, Bertaccini A, Tosi L, Giacopini A, Mazzucchi U, Favaron F, Sella L, Bertaiola F (2015) Vine decline in kiwifruit: climate change and effect on waterlogging and Phytophthora in North Italy. Acta Hortic:93–97
Tacconi G, Giacopini A, Vittone G, Nari L, Spadaro D, Savian F, Ermacora P, Saro S, Morone C, Bardi L, et al. (2019) “Moria del kiwi”: situazione disastrosa al nord, preoccupante nel resto d’Italia. Kiwi informa Aprile/Giugno: 32–37
Tarquini G, Zaina G, Ermacora P, De Amicis F, Franco-Orozco B, Loi N, Martini M, Bianchi GL, Pagliari L, Firrao G et al (2019) Agroinoculation of Grapevine Pinot Gris Virus in tobacco and grapevine provides insights on viral pathogenesis. PLoS One 14:e0214010
Team RC. 2013. R: a language and environment for statistical computing
Tosi L, Tacconi G, Giacopini A (2015) La moria del kiwi, situazione e prospettive. L’Informatore Agrario 44:67–70
Toussoun TA, Bega RV, Nelson PE (1970) Root diseases and soil-borne pathogens. University of California Press, Berkeley
Wallace HR (1978) The diagnosis of plant diseases of complex etiology. Annu Rev Phytopathol 16:379–402
Wang KX, Xie YL, Yuan GQ, Li QQ, Lin W (2015) First report of root and collar rot caused by Phytopythium helicoides on kiwifruit (Actinidia chinensis). Plant Dis 99:725
White TJ, Bruns T, Lee S, Taylor JL (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18:315–322
Xiloyannis C, Massai R, Piccotino D, Baroni G, Bovo M (1993) Method and technique of irrigation in relation to root system characteristics in fruit growing. Acta Hortic:505–510
Xiloyannis C, Nuzzo B, Dichio V, Celano G (1997) L’importanza delle caratteristiche dell’apparato radicale e dell’architettura della chioma nella scelta e nella gestione del metodo irriguo per l’actinidia. Rivista di Frutticoltura 59:31–37
Acknowledgments
Dr. Simone Saro, from ERSA- Phytosanitary Service of Friuli Venezia Giulia region (Italy), for his collaboration and support in field-scouting activities. Mr. Borselli Stefano and Mr. Alberto Loschi from University of Udine, for their professional advice and help when setting up the experiment and throughout the entire duration of the trial. Mr. Arrigo Toffolutti, from Toffolutti nursery, for supplying kiwifruit plantlets.
Funding
Open access funding provided by Università degli Studi di Udine within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Contributions
All authors contributed to the experimental design and the writing of this paper, and more specifically: statistical analysis of rainfall series FG; interpretation of rainfall analysis FS, FG; TEM sample preparation, observation and interpretation FS, LP, RM; Isolation and identification of isolates FS, GT, MM, NS; Interpretation of greenhouse trial FS, PE, RM, MM, GF.
Corresponding author
Additional information
Responsible Editor: Stéphane Compant.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Savian, F., Ginaldi, F., Musetti, R. et al. Studies on the aetiology of kiwifruit decline: interaction between soil-borne pathogens and waterlogging. Plant Soil 456, 113–128 (2020). https://doi.org/10.1007/s11104-020-04671-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-020-04671-5