Abstract
Late blight is a disease with the biggest economic impact on potato cultivation worldwide. Pyramiding of the resistance genes originating from potato wild relatives is a breeding strategy that has a potential to produce potato cultivars durably resistant to late blight. Growing such cultivars would allow limiting the intensive chemical control of the disease. The goal of this work was to transfer the late blight resistance gene Rpi-rzc1 from Solanum ruiz-ceballosii to the tetraploid level of cultivated potato and to pyramid it with the Rpi-phu1 gene. We obtained two diploid and, through 4x-2x cross, a tetraploid potato population segregating for the Rpi-rzc1 presence, as well as one diploid and one tetraploid population where both genes were introgressed. In total, 754 progeny clones were tested for resistance to late blight in detached leaflet assays. Pathogen isolates avirulent on plants with both genes and virulent on plants with the Rpi-phu1 were used. The selection was assisted by two PCR markers flanking the Rpi-rzc1 gene and a newly designed, highly specific intragenic marker indicating the Rpi-phu1 gene presence. We obtained 26 diploid and 49 tetraploid potato clones with pyramid of both genes that should enhance the durability and spectrum of their late blight resistance and that can be exploited in potato breeding. The specificity of the marker for the Rpi-phu1 gene and the precision of the Rpi-rzc1 mapping were improved in this work.
Similar content being viewed by others
Introduction
The cultivated potato, Solanum tuberosum L., and its wild relatives originate from South and Central America. Over 200 species of Solanum section Petota, to which S. tuberosum belongs, include species of various ploidy levels (diploids, tetraploids, hexaploids). Their taxonomy is affected by introgressions, interspecific hybridisations, sexual compatibility and phenotypic plasticity. They show diversity in plant height, tuber, leaf and flower colours and shapes (Ovchinnikova et al. 2011; Spooner et al. 2004). Wild potato relatives are adapted to plethora of environments that involve different precipitation regimes, day lengths, soils or latitudes and altitudes (Machida-Hirano 2015). They are used in potato breeding programs as sources of traits valuable from the breeder’s and consumer’s perspective, with resistance to biotic and abiotic stresses in particular.
Among the biotic stresses, the late blight caused by Phytophthora infestans (Mont.) de Bary is responsible for financial losses associated with yield decrease and cost of fungicides used to control the disease. Starting with a discovery of first 11 late blight resistance (Rpi) genes in S. demissum in 1950s, the number rose currently to over 60 identified genes from at least 25 Solanum species. All Rpi genes cloned so far contain nucleotide-binding site and leucine-rich repeats domains (Rodewald and Trognitz 2013). These domains were found in the reference potato genome in 755 genes distributed over 12 chromosomes in 92 gene clusters (Jupe et al. 2013). Because of the pathogen’s ability for rapid adaptation, introduced Rpi genes may turn ineffective. As a promising strategy to extend the durability of resistance against late blight, gene stacking in a single potato cultivar was proposed. Introgressed Rpi genes should preferably originate from different gene clusters, thus differing in recognition specificity of pathogen gene products (Zhu et al. 2012). However, it was reported that the recognition spectrum of Rpi genes from different gene clusters might overlap, as it was proved for products of the R2 and Rpi-mcq1 genes recognising the same P. infestans protein (Aguilera-Galvez et al. 2018).
The late blight resistance genes were selected for pyramiding in this work because of the wide spectra of provided resistance, none or limited presence in cultivars so far, and locations on different potato chromosomes facilitating selection. The late blight Rpi-rzc1 gene, originating from the diploid species S. ruiz-ceballosii, has been mapped to the chromosome X (Śliwka et al. 2012). The gene provides high level of resistance that has been rarely defeated by P. infestans isolates (Brylińska et al. 2016). The sequence of the Rpi-rzc1 gene is not known. The sequence of the other Rpi gene used in this work, the Rpi-phu1, originating from diploid S. phureja and located at potato chromosome IX (Śliwka et al. 2006), is identical to the Rpi-vnt1.1 gene from S. venturii (Foster et al. 2009). Also the Rpi-phu1 gene provided high resistance against wide spectrum of P. infestans (Brylińska et al. 2016).
So far, no cultivar with the Rpi-rzc1 gene is commercially available. Cultivars carrying the Rpi-vnt1.1 / Rpi-phu1 gene, like Polish cv. Gardena (registration in 2018) or Dutch cv. Alouette (registration in 2014), have already been released, which may exert selection pressure on pathogen population against that gene. Gene stacking might however slower down the process. Potato cultivars containing multiple Rpi genes maintain durable resistance of broader spectrum as reported for cvs. Bzura (R2-like + unidentified Rpi gene(s)), Sárpo Mira (R3a, R3b, R4, R8 and Rpi-Smira1) or Mastenbroek’s differentials R8 (R3a, R3b, R4 and R8) and R9 (R1, Rpi-abpt1, R3a, R3b, R4, R8, R9) that possess at least two to seven known Rpi genes (Plich et al. 2015; Rietman et al. 2012; Kim et al. 2012). Rapid and efficient introduction of several Rpi genes into a single potato line was successfully applied by genetic engineering as done by Zhu et al. (2012), Jo et al. (2014) or most recently by Ghislain et al. (2019). The obtained genotypes exhibited broad-spectrum resistance to late blight and are being developed into registered potato cultivars.
The success of an Rpi gene introduction can be investigated by screening for resistance in phytopathological tests in laboratory or field conditions. However, such tests are laborious and time-consuming. Phenotypical tests can be replaced with molecular markers, which, in marker-assisted selection (MAS), speed up the process of selecting plants carrying the gene of interest.
The aim of this work was to transfer the late blight resistance gene Rpi-rzc1 to the tetraploid level of cultivated potato and to pyramid it with the Rpi-phu1 gene. For selection, besides phenotypical screening, we used molecular markers that flank the Rpi-rzc1 gene and a marker derived from the Rpi-phu1 gene sequence.
Materials and methods
Plant material
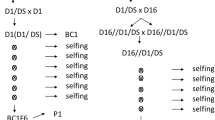
The plant material consisted of five unselected F1 potato progenies: three diploid (G2P-1, G2P-2 and G2P-3) and two tetraploid (4xG-1 and 4xG-2). The parents and numbers of individuals in the populations are presented in the Fig. 1. The crosses were performed between donors of the Rpi-rzc1 gene and potato clones susceptible to late blight (populations G2P-1, G2P-3 and 4xG-1) or between donors of the Rpi-rzc1 gene and donors of the Rpi-phu1 gene (populations G2P-2 and 4xG-2). Pedigrees of the plant material are presented in Fig. 1a (diploids) and Fig. 1b (tetraploids). Origins of the genes Rpi-rzc1 and Rpi-phu1 were described by Śliwka et al. (2012) and Śliwka et al. (2006), respectively. Diploid late blight susceptible parents carried other useful traits: DG 97-2174 and DG 97-943 were suitable for chips production; DG 06-508 was resistant to pectinolytic bacteria and Synchytrium endobioticum.
Pedigree of the material in which the Rpi-rzc1 gene was introduced into cultivated potato breeding-pool and combined with Rpi-phu1 gene, at diploid (a) and tetraploid (b) level. Unselected populations segregating for Rpi-rzc1 or for Rpi-rzc1 and Rpi-phu1 genes are marked by rectangles with thick borders and number of individuals given. Grey rectangles mark diploid potato clones DG 08–238 used as donor of Rpi-rzc1 gene at diploid level and to transfer this gene into tetraploid potato breeding-pool. White rectangles with thin borders show potato breeding lines and cultivars. Resistant clones are marked by the names of resistance genes they were carrying; other clones were susceptible to late blight
Diploid progenies
2x-2x crossing programs were performed according to method described by Jakuczun and Wasilewicz-Flis (2001). Plants used as mother plants were grafted on tomato in order to extend the time of flowering. Flowers were emasculated at the bud stage 1 day before the pollination. Fresh pollen was collected from flowers of pollen parents and stored at about − 20 °C. The pollen fertility was assessed by the staining with lactofuchsin and observation of pollen grains at × 250 magnification, as described by Wasilewicz-Flis and Jakuczun (2001a). The obtained fruits were allowed to mature and then the seeds were extracted. In the spring, the seeds were sown and the seedlings were grown in greenhouse in pots. After plant senescence, tubers of each genotype were harvested separately and stored at about 4–5 °C to the next planting season.
Tetraploid progenies
To introduce the Rpi-rzc1 gene into tetraploid potato germplasm, interploid (4x-2x) and then tetraploid (4x-4x) crosses were performed. In interploid crosses, a diploid potato hybrid clone DG 08-238, producing unreduced (2n) male gametes, was used as a pollen parent. Its ability to produce male 2n gametes was assessed based on the presence of big pollen grains, as described by Wasilewicz-Flis and Jakuczun (2001b). The tetraploid seed parent was a Polish cultivar Tajfun registered in 2004. A catalogue score of field late blight resistance of cv. Tajfun is 5 (in 1–9 scale, where 9 = the most resistant) but in our detached leaflet tests, this cultivar was susceptible (mean score 3.2, P. infestans isolate MP324). In order to extend the time of flowering, tetraploid mother plants were grafted on black nightshade plants (Solanum nigrum L.). Flowers were emasculated at the bud stage just before the pollination. The seeds were sown next spring and the seedlings were grown in greenhouse conditions. The ploidy level of these individuals was confirmed by counting of chloroplasts in guard cells, as described in Wasilewicz-Flis and Jakuczun (2001c). A 4x progeny clone G4-3-5 was chosen for next 4x-4x crosses based on phenotyping results and presence of DNA markers flanking the Rpi-rzc1 gene.
Potato differentials
With each late blight resistance test on dates shown in the Table 1, we performed simultaneously a virulence test against the Black’s differential set (Black et al. 1953) obtained from Scottish Agricultural Science Agency, Edinburgh, UK, and following potato genotypes: cv. Bzura with the R2-like gene (Plich et al. 2015), cv. Sárpo Mira with the R3a, R3b, R4, Rpi-Smira1 and Rpi-Smira2 genes (Rietman et al. 2012; Tomczyńska et al. 2014), cv. Biogold with the Rpi-abpt gene (Park et al. 2005), cv. Toluca with the Rpi-blb2 gene (Zhu et al. 2015), breeding lines 04-IX-21 with the Rpi-phu1 (Śliwka et al. 2013) and 99-10/36 with Rpi-rzc1 genes (Śliwka et al. 2012); the isolate MP324x was additionally tested on cvs. Alouette (Armstrong et al. 2019) and Gardena known to contain Rpi-phu1 / Rpi-vnt1.1 gene (Potato Breeding Zamarte Ltd., Poland, personal communication).
Phenotyping
Detached leaflet assays (DLA) for late blight resistance were performed on fully expanded lateral leaflets collected from the 6-week-old plants as described by Brylińska and Śliwka (2017). Three P. infestans isolates were used (Table 2): MP324 used previously to identify the Rpi-phu1 and Rpi-rzc1 genes (Śliwka et al. 2006; Śliwka et al. 2012), a recent isolate MP1820 representing the 13_A2 (Blue-13) SSR genotype and MP324x virulent on plants with the Rpi-phu1 gene (Stefańczyk et al. 2017). The assays were conducted in 2016–2019 according to the scheme shown in Table 1, one test each year with the exception of 2018 when two independent tests were done on different dates. Each test consisted of two replications with three leaflets of each potato individual per replication. The inoculum was prepared as described by Sobkowiak and Śliwka (2017) and adjusted to the concentration of 50 sporangia μl−1. Leaflets inoculated with a pathogen sporangia suspension were incubated 1 day at 16 °C/no light/abaxial side up, followed by 5 days at 16 °C/1600 lx light/adaxial side-up conditions. Scoring was done after 6 days on a 1–9 scale with 1 meaning leaflet being completely diseased with intensive sporulation visible (Brylińska and Śliwka 2017; Online Resource 1).
Marker-assisted selection
DNA was isolated from young leaves of all potato progeny individuals and parental forms, frozen in liquid nitrogen, using the DNeasy Plant Mini kit (QIAGEN Polska Sp. z o. o., Poland) according to the producer’s instructions. Three PCR markers were used for verification of the Rpi-phu1 and Rpi-rzc1 gene presence. The phu1_2069 primers were designed based on the sequence of the Rpi-vnt1.1 gene (NCBI GenBank: FJ423044) that is identical to the Rpi-phu1 gene (Foster et al. 2009). Primer pair (forward: 5′-CCAAATTACTTGATCATGATT-3′, reverse: 5′-TAGTACCTGTGATATTCTCA-3′) yielded a product that spanned 77% (2069 of 2680 bp) of the Rpi-phu1 gene. PCR mixture (20 μL) contained sterile water, 1 μL of each 10 μM primer solution, 2 μL of 2 mM dNTPs, 4 μL of 5× high fidelity buffer and 0.4 U of Phusion DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, US). A touchdown PCR technique was used with initial denaturation for 180 s at 95 °C, followed by 7, 8 and 20 cycles of 95 °C for 30 s, 67/65/62 °C for 20 s and 72 °C for 45 s, ending with final elongation of 72 °C for 300 s. RenSeq1812 and RenSeq1910 primer sets were used for amplifying, respectively, 520 and 350 bp markers flanking the Rpi-rzc1, each in a distance of 0.4 cM (Brylińska et al. 2015). PCR mixture contained 0.4 μL of each 10 μM primer solution, 1 μL of 2 mM dNTPs, 2 μL of 10× PCR buffer and 1 U of Taq Polymerase (Genoplast Chemicals, Rokocin, Poland), filled up with water to 20 μL. Both markers were amplified using the following PCR program: 180 s at 95 °C, 30 cycles of 94 °C for 30 s, 55 °C for 45 s and 72 °C for 45 s, finished with 420 s at 72 °C. The products obtained with the RenSeq1812 and RenSeq1910 primers were digested with, respectively, BsaJI and HinfI restriction endonucleases. For this purpose, 10 μL of PCR product was mixed with 3 U of restriction enzyme, 2 μL of 10× corresponding buffer and filled up with sterile water to 20 μL. Mixtures were then incubated for 3 h at 55 °C (BsaJI) or 37 °C (HinfI). The PCR and digestion products were separated on 1.5% agarose gels stained with ethidium bromide and visualised using the UV light. Appearance of a 500 bp and/or 290 bp bands after digestion of the RenSeq1812 and RenSeq1910 PCR products indicated the presence of the Rpi-rzc1 gene. The DNA samples of the potato tetraploid genotypes Z-03.3827 (Stefańczyk et al. 2017), 04-IX-21, cvs. Gardena and Alouette were used as the controls of the PCR assays carrying the Rpi-phu1 gene, and of a diploid clone DG 99-10/36 (Śliwka et al. 2012) as a carrier of the Rpi-rzc1 gene. DNAs of the late blight resistant cvs. Carolus and Kelly were used as negative controls in the phu1_2069 marker amplification.
Statistical analyses
All the statistical analyses were performed using Statistica 10 software (Statsoft Inc. 2011). To test for the deviation between observed and expected segregation ratios, the chi-square (χ2) test was applied. The correlations between the results of particular DLA done in different years and with the use of P. infestans isolates were assessed by calculation of Pearson’s correlation coefficients. Analysis of variance and post-hoc Tukey’s range test for unequal sample sizes were applied to study the effects of potato genotype and P. infestans isolate on late blight resistance scores.
Results
Crosses
The pollen fertility of DG 99-10/36, the donor of the Rpi-rzc1 gene, was assessed as 50%, and no big pollen grains (2n male gametes) were observed. This clone was crossed as pollen parent with two diploid clones DG 97-2174 and DG 97-943 (Fig. 1), and the efficiency of these pollinations was high. In the case of DG 97-2174 × DG 99-10/36 crosses, 36 flowers were pollinated and 22 berries were obtained. From these berries, we extracted in total 2594 seeds. In the case of DG 97-943 × DG 99-10/36 crosses, 22 flowers were pollinated and 9 berries were obtained. From these berries, 1112 seeds were extracted.
The Rpi-rzc1-containing diploid potato clone DG 08-238 was used as pollen parent in 2x-2x crosses as well as in 4x-2x crosses, to transfer the Rpi-rzc1 gene from diploid to tetraploid potato gene-pool. The diploid level of DG 08-238 was confirmed, and the mean number of chloroplasts in guard cells was 8.6. The fertility of its pollen varied from 40 to 70%. The presence of big pollen grains (2n gametes) in this clone was confirmed. From 282 flowers of diploid clone DG 06-508 pollinated with the DG 08-238 pollen, we obtained 43 berries and 1335 seeds. The efficiency of pollination in interploid 4x-2x crosses was much lower. From 122 pollinated flowers of tetraploid cv. Tajfun, we obtained 19 berries and only 14 seeds, of which 12 plants emerged. The Rpi-rzc1-containing progeny clone G4-3-5, obtained from 4x-2x cross, was tetraploid with the mean number of chloroplasts in guard cells 22.9. This clone was used as seed parent in 4x-4x crosses with tetraploid potato cultivar Felka Bona, and with Rpi-phu1-containing tetraploid clone Bio-7. The effectiveness of these crosses was very high: 29 berries and 1280 seeds were obtained from cross G4-3-5 × Bio-7, while from cross G4-3-5 × Felka Bona, 47 berries and 4300 seeds were obtained. The ploidy level of 10 randomly selected clones from each of tetraploid progenies 4xG-1 and 4xG-2 was assessed, and all were tetraploid (range of mean of chloroplasts numbers: 20.8–23.0).
Genotyping
To detect the presence of the Rpi-rzc1 gene, we obtained PCR products with the DNA of all 754 individual plants tested, using the RenSeq1812 and RenSeq1910 markers. After restriction digestion, the presence of the bands of approx. 500 and 290 bp, linked to the Rpi-rzc1 gene, was scored for the RenSeq1812 and RenSeq1910 markers, respectively. There were four recombinants between the two markers, two in a diploid population G2P-1 and two in a tetraploid population 4xG-1. The numbers of RenSeq1910-positive and negative-individuals in each population and the recombinants are shown in Table 3.
In plants of populations G2P-2 and 4xG-2, the amplification of the phu1_2069 marker resulted in a single strong band of 2069 bp in size in the individuals with the Rpi-phu1 gene. As a positive control, the DNA of the 04-IX-21 potato line was used (Fig. 2). Another band of around 1900 bp can be additionally amplified using the marker: both bands were simultaneously obtained with the Z-03.3827 genotype (Stefańczyk et al. 2017) and with the cv. Alouette, while in cvs. Carolus and Kelly, the 1900 bp band was amplified only (Fig. 2). In total, there were 56 and 84 phu1_2069 marker positive individuals in populations G2P-2 and 4xG-2, respectively (Table 3).
Phenotyping
The virulence of the three P. infestans isolates in the DLA, taken as a consensus from historical (Stefańczyk et al. 2017; Śliwka et al. 2013; Tomczyńska et al. 2014) and the data obtained in this study, is summarised in the Table 2. None of the isolates was able to infect the 99-10/36 plants carrying the Rpi-rzc1 gene. The plants with the Rpi-phu1 gene: cvs. Gardena and Alouette, as well as potato lines 04-IX-21 and Z-03.3827, were infected only with the MP324x isolate and showed no symptoms of infection with other isolates used. The isolate MP324 was able to infect cv. Bzura, while cvs. Sárpo Mira, Toluca and Biogold remained moderately resistant. The isolate MP1820 exhibited similar virulence profile to MP324, except it completely infected cv. Biogold, but left cv. Sárpo Mira less affected by the disease.
All the individuals from the five potato populations were phenotyped in the DLA according to the scheme presented in the Table 1. Two P. infestans isolates used in the assays, MP324 and MP1820, were avirulent on plants with the Rpi-phu1 and Rpi-rzc1 genes and of similar virulence profiles (Table 2). The resistance scores in DLA obtained with both isolates were strongly correlated in each of the populations surveyed (Pearson’s correlation coefficients were 0.933, 0.977 and 0.958 at p < 0.001 for populations G2P-1, G2P-2 and G2P-3, respectively). The analysis of variance indicated no significant effect of the isolate on average resistance score (Online Resource 2). The resistance data obtained with the isolates MP324 and MP1820 were therefore pooled together for further analyses (Figs. 3 and 4).
Box plots of the resistance in three potato populations divided into groups with the genetic markers indicating the presence of the Rpi-rzc1 late blight resistance gene (x-axis). The mean values of the resistance scores against the P. infestans isolates MP324 and MP1820 are shown as squares, the box represents standard error, and the whiskers indicate standard deviation. Significant differences were calculated using Tukey’s range test for unequal sample sizes with a significance level of 0.05. When marked with the same letter, groups do not differ significantly
Box plots of the resistance in two potato populations divided into groups with the genetic markers indicating the presence of the Rpi-rzc1 and Rpi-phu1 late blight resistance genes (x-axis). a, c Mean results of tests with P. infestans isolates avirulent on Rpi-phu1 plants, MP324 and MP1820. b, d Mean results of tests with P. infestans isolate MP324x, virulent on Rpi-phu1 plants. The mean values of the resistance against the P. infestans isolates are shown as squares; the box represents standard error, and the whiskers indicate standard deviation. Significant differences were calculated using Tukey’s range test for unequal sample sizes with a significance level of 0.05. When marked with the same letter, groups do not differ significantly
Marker-assisted selection
In each potato population tested, the RenSeq1910 marker co-segregated with the resistance scored in DLA, while four individuals showed recombination between the RenSeq1812 marker and the gene (Table 3). For one recombinant from 4xG-1 population, a product with the RenSeq1812 marker was obtained, while no product was obtained with the RenSeq1910 marker, and DLA results indicated absence of the Rpi-rzc1 gene. The other three recombinants (one from 4xG-1 and two from G2P-1 populations) gave no product with the RenSeq1812, whereas RenSeq1910 marker and phenotype tests indicated the Rpi-rzc1 gene presence. The percentages of recombinants in the G2P-1 and 4xG-1 populations were estimated at 1.87% and 1.10%, respectively.
In two Rpi genes-stacking populations, where the Rpi-phu1 gene segregated, neither false negative nor false positive results were obtained with the phu1_2069 marker when we compared the phenotype and genotype of plants in three groups: without both Rpi genes, with the Rpi-phu1 or with the Rpi-rzc1 only. In the groups with both genes, the presence of the Rpi-phu1 gene was not verified phenotypically due to the lack of P. infestans isolate virulent on the Rpi-rzc1 plants.
Based on the molecular markers and the DLA results, we diagnosed the presence of the Rpi-rzc1 and Rpi-phu1 resistance genes in each of the examined potato population. The numbers of the individual plants containing both investigated Rpi genes (i.e. simultaneously positive for RenSeq1910, RenSeq1812 and phu1_2069 markers) were 26 and 49 in the diploid and tetraploid populations, respectively (Table 3).
The results presented in Figs. 3 and 4 are showing the resistance of plant material against P. infestans isolates avirulent (Fig. 3; Fig. 4a, c), or virulent (Fig. 4b, d), on plants with the Rpi-phu1 gene. The differences in resistance of plants with or without the Rpi-rzc1 gene were significant in all populations and tests (Figs. 3 and 4). Plants with or without the Rpi-phu1 gene differed significantly only in tests with the P. infestans isolates MP324 and MP1820 (Fig. 3; Fig. 4a, c). Individuals without the Rpi-rzc1 gene that belonged to the G2P-1, G2P-3 and 4xG-1 populations did not differ significantly in resistance to late blight according to the Tukey’s test and were scored at 2.4, 2.0 and 1.5 on average, respectively. Individuals from the same populations, but carrying the gene, had average resistance scores estimated at 8.8, 8.9 and 8.9 (Fig. 3), and the difference between these groups was insignificant as well. In the G2P-2 and 4xG-2 populations, regardless of the isolate used, the infected plants without investigated resistance genes were evaluated in DLAs at 1.3–2.4 on average, while the diseased plants with the Rpi-phu1 gene (tests with virulent P. infestans isolate MP324x) had their resistance estimated at 1.8–2.4, and no significant differences were observed between these groups. Similarly, the score of the healthy leaflets in each genotype/P. infestans isolate combination was 8.8–9.0 (Fig. 4) and did not differ significantly between the healthy individuals with the Rpi-phu1, with the Rpi-rzc1 or with both genes present.
The results of the χ2 test for 1:1 and 1:1:1:1 of one or two resistance genes segregation ratios, based on both DNA marker data and DLA results, are presented in Table 3. The assumed 1:1 segregation was confirmed for the G2P-1 and 4xG-1 potato populations, but rejected for G2P-3 in which a deficiency of individuals carrying the Rpi-rzc1 gene was observed. According to the test, the numbers of individuals in four genotype groups did not fit to 1:1:1:1 ratio neither in the G2P-2 nor 4xG-2 populations (Table 3). In the G2P-2 potato population, an excessive amount of plants with no investigated genes, and with the Rpi-rzc1 gene only, was observed. In the 4xG-2 population, there were too many individuals with the Rpi-rzc1 and less than expected plants with the Rpi-phu1 gene.
Discussion
Resistance genes stacking in a single cultivar should improve durability of the resistance, because it is less likely for a pathogen to simultaneously gain virulence towards a plant with several R genes. Gene stacking can be conducted by pyramiding R genes, different alleles of the same gene or even the same alleles in order to observe gene allele-dosage effect (Tan et al. 2010). In some plant species, cultivars with multiple R genes against various biotic stresses have shown durable resistance, e.g. in wheat against Puccinia triticina and P. striiformis for over 20 years each (Mallard et al. 2005; McCallum et al. 2016), in rice against Magnaporthe oryzae “throughout a century of cultivation “(Fukuoka et al. 2015) or in oilseed rape against Leptosphaeria maculans for at least 5years (Brun et al. 2009).
This strategy is used also in potato breeding for some time now, as cultivars such as Pentland Dell from 1960s and Escort from 1980s, but also already the R8 or R9 Black’s differentials from 1950s, are known to contain multiple Rpi genes (Bormann et al. 2004; Tan et al. 2010). Some other promising genotypes, with multiple Rpi genes, are those obtained by Tan et al. (2010), Ghislain et al. (2019) or potato cvs. Bzura and Sárpo Mira (Plich et al. 2015; Rietman et al. 2012). The resistance of cvs. Pentland Dell and Escort turned ineffective soon after registration of these cultivars. The durability of resistance of the R8 and R9 Black’s differentials has never been properly tested as they have never been cultivated on a large scale. These differentials still maintain high resistance against late blight (Brylińska et al. 2016), just as the late maturing cv. Sárpo Mira (Rietman et al. 2012) registered in 2002 and cultivated on moderate scale. Limited success of gene pyramids against potato late blight can be at least partially associated with P. infestans biology and partially with the choice of the Rpi genes used (Leesutthiphonchai et al. 2018). This pathogen is attributed with features that are known to accelerate adaptation and thus R gene pyramid defeat: its genome undergoes frequent modifications through mutations, the pathogen can reproduce sexually, its populations are large and not isolated which allows an increased gene flow (Stam and McDonald 2018). Ideally, Rpi genes used for stacking should not be previously present in any plant cultivar, so adapted isolates would not exist at all or exist in a pathogen population at low frequencies, but that’s rarely a case. To efficiently fight P. infestans, the strategy of using potato cultivars with stacked resistance genes should still be complemented with other management practices (Stam and McDonald 2018).
To respond to fast evolution of a pathogen, a breeder should have access to wide pool of R genes and tools for their fast introduction to breeding lines. Genetic engineering would extend possibilities of gene pyramiding as it allows transferring gene of interest from any source, regardless of crossing barriers, into a crop plant (Mekonnen et al. 2017). Recent advances in development of homozygous potato inbred lines are also promising and provide another approach for generating late blight resistant diploid hybrids. This strategy was used by Su et al. (2020) who obtained potato lines with pyramids of two Rpi genes in different combinations from four wild relatives: S. avilesii, S. chacoense, S. tarijense and S. venturii.
Stacking several resistance genes in a single genotype is challenging. Identifying plants with two or more genes solely by phenotyping can be impossible, if pathogen isolates able to overcome the resistance provided by each gene are not available. If DNA markers are available, whether linked to or based on a gene sequence, a resistance gene presence can be determined with ease. DNA markers solve also the difficulty of phenotypic tests being most often destructive for plants.
In this paper, the Rpi-rzc1 gene was transferred from diploid to the tetraploid level and combined with the Rpi-phu1 gene both in 2x and 4x forms. Individual plants from the obtained 2x and 4x populations were tested with P. infestans isolates, one of which was capable of infecting plants with the Rpi-phu1 gene. We did not possess an isolate defeating Rpi-rzc1-conferred resistance, yet using a molecular marker based on the sequence of the Rpi-phu1 gene asserted reliability of the genotyping approach. On the other hand, the sequence of the Rpi-rzc1 gene is not known and genotyping was performed using two linked markers. As demonstrated in this study, they can be successfully used on both, di- and tetraploid levels. Pre-breeding is often performed on diploid level in potato, especially for introgression of wild germplasm and combining desired traits from different sources. Transfer of genes and DNA markers between ploidy levels may not be straightforward and requires validation, such as in this study, due to the effect of tetraploid cultivars’ genetic background. The obtained breeding lines can be exploited in both di- and tetraploid breeding programs as well as studies on P. infestans–potato interactions.
Physical distance between markers RenSeq1910 (PGSC0003DMG400008595, chr10 52,694,086 bp) and RenSeq1812 (PGSC0003DMG400011045, chr10 53,377,529 bp) in the potato reference genomes DM1–3 is 683,983 bp (Brylińska et al. 2015). Genetic distance in the S. ruiz-ceballosii genome of these two flanking markers was previously estimated at 0.4 cM from the Rpi-rzc1 gene (Brylińska et al. 2015), but this study indicated that RenSeq1910 is located closer to the R gene than the marker RenSeq1812. The distance of the RenSeq1812 marker to the Rpi-rzc1 gene was estimated at 1.87 and 1.10 cM in the G2P-1 and 4xG-1 potato populations, respectively. The RenSeq1910 co-segregated with the Rpi-rzc1 gene in all five tested progenies. In lack of intragenic markers, the linked ones should be closer than 5 cM genetic distance and preferably flanking the gene of interest (Collard and Mackill 2007).
Previous study reported no deviation from the 1:1 segregation ratio of the Rpi-rzc1 gene (Śliwka et al. 2012). However, the results from studies on the Rpi-phu1 gene were depending on the genetic background. In the diploid mapping population 97-30, two -thirds of plants were highly resistant (Śliwka et al. 2006). In three populations obtained later and with higher contribution of S. tuberosum genome, the gene segregated in the 1:1 ratio (Śliwka et al. 2010). In this study, three of five surveyed potato populations exhibited significant deviation from the expected 1:1 and 1:1:1:1 ratios as tested using the χ2 test. This deviation might be caused by a distorted segregation, a phenomenon associated with genetic factors involved in reproduction and observed in plants, also extensively described in potato crosses (Manrique-Carpintero et al. 2016). Distorted segregation may be caused by genetic incompatibilities, but also by skewed sampling during population development, i.e. unaware preferential selection (Boopathi 2013).
In the previous work (Stefańczyk et al. 2017), we have used the phu6 marker to indicate the presence of the Rpi-phu1 gene in breeding lines derived from crosses between the gene donors and cv. Sárpo Mira. The phu6 marker scores were in accordance with the phenotype in 84% cases (Stefańczyk et al. 2017). The disagreement was related to both false negative and false positive results but the latter were more frequent in total. The application of the phu1_2069 marker in this work showed no discrepancy with phenotypes scored in DLA in the genotype groups, in which such comparison was possible. The new marker spanned a larger fragment of the Rpi-phu1 gene and was more specific than the previous phu6.
In this work, we describe successfully developed di- and tetraploid potato populations and selection of 26 and 49 individual plants, respectively, that contain a pyramid of two broad-spectrum late blight resistance genes Rpi-rzc1 and Rpi-phu1. We validated and improved specificity of the PCR markers used for selection that should accelerate and facilitate breeding, but only large-scale cultivation of the future potato varieties can demonstrate the durability of the resistance provided by this gene pyramid.
Availability of data and material
All data are presented in the manuscript. Phytophthora infestans isolates are available from Młochów Potato Pathogen Collection and selected potato breeding lines from Młochów Potato Collection at Plant Breeding and Acclimatization Institute–National Research Institute, Poland.
References
Aguilera-Galvez C, Champouret N, Rietman H, Lin X, Wouters D, Chu Z, Jones JDG, Vossen JH, Visser RGF, Wolters PJ, Vleeshouwers VGAA (2018) Two different R gene loci co-evolved with Avr2 of Phytophthora infestans and confer distinct resistance specificities in potato. Stud Mycol 89:105–115
Armstrong MR, Vossen J, Lim TY, Hutten RCB, Xu J, Strachan SM, Harrower B, Champouret N, Gilroy EM, Hein I (2019) Tracking disease resistance deployment in potato breeding by enrichment sequencing. Plant Biotechnol J 17:540–549
Black W, Mastenbroek C, Mills WR, Peterson LC (1953) A proposal for an international nomenclature of races of Phytophthora infestans and of genes controlling immunity in Solanum demissum derivatives. Euphytica 2:173–179
Boopathi NM (2013) Genetic mapping and marker assisted selection. Springer, New York
Bormann CA, Rickert AM, Castillo Ruiz RA, Paal J, Lübeck J, Strahwald J, Buhr K, Gebhardt C (2004) Tagging quantitative trait loci for maturity-corrected late blight resistance in tetraploid potato with PCR-based candidate gene markers. Mol Plant-Microbe Interact 17:1126–1138
Brun H, Chèvre AM, Fitt BDL, Powers S, Besnard AL, Ermel M, Huteau V, Marquer B, Eber F, Renard M, Andrivon D (2009) Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus. New Phytol 185:285–299
Brylińska M, Śliwka J (2017) Laboratory assessment of potato resistance to Phytophthora infestans. Plant Breeding and Seed Science 76:17–23
Brylińska M, Tomczyńska I, Jakuczun H, Wasilewicz-Flis I, Witek K, Jones JDG, Śliwka J (2015) Fine mapping of the Rpi-rzc1 gene conferring broad-spectrum resistance to potato late blight. Eur J Plant Pathol 143:193–198
Brylińska M, Sobkowiak S, Stefańczyk E, Śliwka J (2016) Potato cultivation system affects population structure of Phytophthora infestans. Fungal Ecol 20:132–143
Collard BCY, Mackill DJ (2007) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc B 363:557–572
Foster SJ, Park TH, Pel M, Brigneti G, Śliwka J, Jagger L, van der Vossen E, Jones JDG (2009) Rpi-vnt1.1, a Tm-2(2) homolog from Solanum venturii, confers resistance to potato late blight. Mol Plant-Microbe Interact 22:589–600
Fukuoka S, Saka N, Mizukami Y, Koga H, Yamanouchi U, Yoshioka Y, Hayashi N, Ebana K, Mizobuchi R, Yano M (2015) Gene pyramiding enhances durable blast disease resistance in rice. Sci Rep 5:7773
Ghislain M, Byarugaba AA, Magembe E, Njoroge A, Rivera C, Román ML, Tovar JC, Gamboa S, Forbes GA, Kreuze JF, Barekye A, Kiggundu A (2019) Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnol J 17(6):1119–1129
Jakuczun H, Wasilewicz-Flis I (2001) Production of true potato seeds. Plant Breeding and Acclimatization Institute, Radzików, Poland. IHAR Monografie i Rozprawy Naukowe 10a: 118-20
Jo KR, Kim CJ, Kim SJ, Kim TY, Bergervoet M, Jongsma MA, Visser RGF, Jacobsen E, Vossen JH (2014) Development of late blight resistant potatoes by cisgene stacking. BMC Biotechnol 14:50
Jupe F, Witek K, Verweij W, Śliwka J, Pritchard L, Etherington GJ, Maclean D, Cock PJ, Leggett RM, Bryan GJ, Cardle L, Hein I, Jones JD (2013) Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J 76:530–544
Kim HJ, Lee HR, Jo KR, Mortazavian SM, Huigen DJ, Evenhuis B, Kessel G, Visser RG, Jacobsen E, Vossen JH (2012) Broad spectrum late blight resistance in potato differential set plants MaR8 and MaR9 is conferred by multiple stacked R genes. Theor Appl Genet 124:923–935
Leesutthiphonchai W, Vu AL, Ah-Fong AMV, Judelson HS (2018) How does Phytophthora infestans evade control efforts? Modern insight into the late blight disease. Phytopathology 108:916–924
Machida-Hirano R (2015) Diversity of potato genetic resources. Breed Sci 65:26–40
Mallard S, Gaudet D, Aldeia A, Abelard C, Besnard AL, Sourdille P, Dedryver F (2005) Genetic analysis of durable resistance to yellow rust in bread wheat. Theor Appl Genet 110:1401–1409
Manrique-Carpintero NC, Coombs JJ, Veilleux RE, Buell CR, Douches DS (2016) Comparative analysis of regions with distorted segregation in three diploid populations of potato. G3: genes. Genomes, Genetics 6:2617–2628. https://doi.org/10.1534/g3.116.030031
McCallum BD, Hiebert CW, Cloutier S, Bakkeren G, Rosa S, Humphreys D, Marias G, McCartney C, Panwar V, Rampitsch C, Saville B, Wang X (2016) A review of wheat leaf rust research and the development of resistant cultivars in Canada. Can J Plant Pathol 38:1–18
Mekonnen T, Haileselassie T, Tesfaye K (2017) Identification, mapping and pyramiding of genes/quantitative trait loci (QTLs) for durable resistance of crops to biotic stresses. Journal of Plant Pathology and Microbiology 8:412. https://doi.org/10.4172/2157-7471.1000412
Ovchinnikova A, Krylova E, Gavrilenko T, Smekalova T, Zhuk M, Knapp S, Spooner DM (2011) Taxonomy of cultivated potatoes (Solanum section Petota: Solanaceae). Bot J Linn Soc 165:107–155
Park TH, Vleeshouwers VG, Hutten RCB (2005) High-resolution mapping and analysis of the resistance locus Rpi-abpt against Phytophthora infestans in potato. Mol Breed 16:33–43
Plich J, Tatarowska B, Lebecka R, Śliwka J, Zimnoch-Guzowska E, Flis B (2015) R2-like gene contributes to resistance to Phytophthora infestans in polish potato cultivar Bzura. Am J Potato Res 92:350–358
Rietman H, Bijsterbosch G, Cano LM, Lee HR, Vossen JH, Jacobsen E, Visser RG, Kamoun S, Vleeshouwers VG (2012) Qualitative and quantitative late blight resistance in the potato cultivar Sárpo Mira is determined by the perception of five distinct RXLR effectors. Mol Plant-Microbe Interact 25:910–919
Rodewald J, Trognitz B (2013) Solanum resistance genes against Phytophthora infestans and their corresponding avirulence genes. Mol Plant Pathol 14:740–757
Śliwka J, Jakuczun H, Lebecka R, Marczewski W, Gebhardt C, Zimnoch-Guzowska E (2006) The novel, major locus Rpi-phu1 for late blight resistance maps to potato chromosome IX and is not correlated with long vegetation period. Theor Appl Genet 113:685–695
Śliwka J, Jakuczun H, Kamiński P, Zimnoch-Guzowska E (2010) Marker-assisted selection of diploid and tetraploid potatoes carrying Rpi-phu1, a major gene for resistance to Phytophthora infestans. J Appl Genet 51:133–140
Śliwka J, Jakuczun H, Chmielarz M, Hara-Skrzypiec A, Tomczyńska I, Kilian A, Zimnoch-Guzowska E (2012) Late blight resistance gene from Solanum ruiz-ceballosii is located on potato chromosome X and linked to violet flower colour. BMC Genet 13:11
Śliwka J, Świątek M, Tomczyńska I, Stefańczyk E, Chmielarz M, Zimnoch-Guzowska E (2013) Influence of genetic background and plant age on expression of the potato late blight resistance gene Rpi-phu1 during incompatible interactions with Phytophthora infestans. Plant Pathol 62:1072–1080
Sobkowiak S, Śliwka J (2017) Phytophthora infestans: isolation of pure cultures, storage and inoculum preparation. Plant Breeding and Seed Science 76:9–15
Spooner DM, van den Berg RG, Rodríguez A, Bamberg J, Hijmans RJ, Cabrera SIL (2004) Wild potatoes (Solanum section Petota; Solanaceae) of North and Central America. Syst Bot Monogr 68:1–209
Stam R, McDonald BA (2018) When resistance gene pyramids are not durable – the role of pathogen diversity. Mol Plant Pathol 19:521–524
StatSoft Inc (2011) STATISTICA (data analysis software system), version 10. Available from: www.statsoft.com
Stefańczyk E, Sobkowiak S, Brylińska M, Śliwka J (2017) Expression of the potato late blight resistance gene Rpi-phu1 and Phytophthora infestans effectors in the compatible and incompatible interactions in potato. Phytopathology 107:740–748
Su Y, Viquez-Zamora M, den Uil D, Sinnige J, Kruyt H, Vossen J, Lindhout P, van Heusden S (2020) Introgression of genes for resistance against Phytophthora infestans in diploid potato. Am J Potato Res 97:33–42
Tan MYA, Hutten RCB, Visser GFV, van Eck HJ (2010) The effect of pyramiding Phytophthora infestans resistance genes RPi-mcd1 and RPi-ber in potato. Theor Appl Genet 121:117–125
Tomczyńska I, Stefańczyk E, Chmielarz M, Karasiewicz B, Kamiński P, Jones JDG, Lees A, Śliwka J (2014) A locus conferring effective late blight resistance in potato cultivar Sárpo Mira maps to chromosome XI. Theor Appl Genet 127:647–657
Wasilewicz-Flis I, Jakuczun H (2001a) Estimation of pollen fertility. Plant Breeding and Acclimatization Institute, Radzików, Poland. IHAR Monografie i Rozprawy Naukowe 10a: 121-2
Wasilewicz-Flis I, Jakuczun H (2001b) Evaluation of the ability to produce male unreduced gametes (2n) in diploid potatoes. Plant Breeding and Acclimatization Institute, Radzików, Poland. IHAR Monografie i Rozprawy Naukowe 10a:126–127
Wasilewicz-Flis I, Jakuczun H (2001c) Estimation of ploidy level in potato. Plant Breeding and Acclimatization Institute, Radzików, Poland. IHAR Monografie i Rozprawy Naukowe 10a: 123-5
Zhu S, Li Y, Vossen JH, Visser RGF, Jacobsen E (2012) Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Res 21:89–99
Zhu S, Vossen JH, Bergervoet M, Nijenhuis M, Kodde L, Kessel GJT, Vleeshouwers VG, Visser RGF, Jacobsen E (2015) An updated conventional- and a novel GM potato late blight R gene differential set for virulence monitoring of Phytophthora infestans. Euphytica 202:219–234
Acknowledgements
We thank Professor Ewa Zimnoch-Guzowska (Plant Breeding and Acclimatization Institute–National Research Institute, Poland) for critical comments on the manuscript and Mrs. Małgorzata Frączak and Anna Jarzyńska for technical assistance and DNA extraction in particular.
Funding
The research was financed within G2P-SOL project (Title: Linking genetic resources, genomes and phenotypes of Solanaceous crops) that has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 677379.
Author information
Authors and Affiliations
Contributions
ES optimised and applied PCR markers. JP performed some of the crosses and maintained the plant material and collected plant material for DNA extraction. SS maintained P. infestans cultures and prepared inocula for detached leaflet tests. MJ and JP performed detached leaflet tests. ES, JP and JS were involved in data analyses and manuscript writing. MJ, PSD and SS proofread the manuscript. JS contributed to designing the research.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stefańczyk, E., Plich, J., Janiszewska, M. et al. Marker-assisted pyramiding of potato late blight resistance genes Rpi-rzc1 and Rpi-phu1 on di- and tetraploid levels. Mol Breeding 40, 89 (2020). https://doi.org/10.1007/s11032-020-01169-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-020-01169-x