Abstract
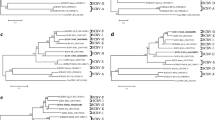
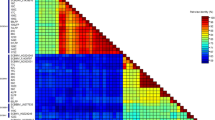
Banana streak virus (BSV) is the causal agent of the most prevalent diseases in banana and causes serious economic losses to banana production worldwide. To examine variability in BSV, 65 isolates were obtained from 99 samples collected from Guangdong, Hainan, Guangxi and Yunnan provinces in South China. The 65 BSV isolates shared 62.8–97.8% nt identity over the RT/RNase H regions when compared to eight BSV species with published complete genomes. Sequence and phylogenetic analyses based on the RT/RNase H region indicated that there was great genetic diversity among BSV isolates in South China, and some isolates belonged to species of badnavirus that had never before been reported in China, including banana streak OL virus (BSOLV) (DN6–2), banana streak IM virus (BSIMV) (DN1–7 and DN5–1), and banana streak VN virus (BSVNV) (YLJ-8). The full genomes of two isolates (DN5–1and DN5–4) were cloned and sequenced. The genetic characteristics of both isolates were very close to those of typical badnaviruses. Based on percentage nucleotide sequence identity and phylogenetic relationships, DN5–1 was categorized as BSIMV. Interestingly, while DN5–4 was similar to both sugarcane bacilliform viruses (SCBVs) and BSVs based on sequence identities of the RT/RNase H region, analysis of its complete sequence and genome organization revealed that it can be further classified as sugarcane bacilliform Guadeloupe A virus-2 (SCBGAV-2). To our knowledge, this is the first report of natural infection of banana by SCBGAV-2 in China. In addition, our results suggest that considering the complete genomic sequence rather than the RT/RNase H region alone is important when classifying badnavirus isolates.




Similar content being viewed by others
References
Adams MJ, Carstens EB (2012) Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses (2012). Arch Virol 157:1411–1422. https://doi.org/10.1007/s00705-012-1299-6
Bhat AI, Hohn T, Selvarajan R (2016) Badnaviruses: the current global scenario. Viruses 8(6):177. https://doi.org/10.3390/v8060177
Bousalem M, Durand O, Scarcelli N, Lebas BSM, Kenyon L, Marchand JL, Lefort F, Seal SE (2009) Dilemmas caused by endogenous pararetroviruses regarding the taxonomy and diagnosis of yam (Dioscorea spp.) badnaviruses: analyses to support safe germplasm movement. Arch Virol 154:297–314. https://doi.org/10.1007/s00705-009-0311-2
Borah BK, Sharma S, Kant R, Johnson AMA, Saigopal DVR, Dasgupta I (2013) Bacilliform DNA-containing plant viruses in the tropics: commonalities within a genetically diverse group. Mol Plant Pathol 14:759–771. https://doi.org/10.1111/mpp.12046
Bouhida M, Lockhart BE, Olszewski NE (1993) An analysis of the complete sequence of a sugarcane bacilliform virus genome infectious to banana and rice. J Gen Virol, 74(pt 1):15–22. https://doi.org/10.1099/0022-1317-74-1-15
Dahal G, Hughes JDA, Thottappilly G, Lockhart BEL (1998) Effect of temperature on symptom expression and reliability of banana streak badnavirus detection in naturally infected plantain and banana (Musa spp.). Plant Dis 82:16–21. https://doi.org/10.1094/PDIS.1998.82.1.16
Dahal G, Ortiz R, Tenkouano A, D’Hughes JA, Thottappilly G, Vuylsteke D, Lockhart BEL (2000) Relationship between natural occurrence of banana streak badnavirus and symptom expression, relative concentration of viral antigen, and yield characteristics of some micropropagated Musa spp. Plant Pathol 49:68–79(12). https://doi.org/10.1046/j.1365-3059.2000.00420.x
Fei J, Xiao H, Li H, Fan H (2001) Progress of banana streak disease caused by banana streak badnavirus. Chin J Virol 17:381–385. https://doi.org/10.3321/j.issn:1000-8721.2001.04.018
Gawel NJ, Jarret RL (1991) A modified CTAB DNA extraction procedure for Musa and Ipomoea. Plant Mol Biol Report 9:262–266. https://doi.org/10.1007/BF02672076
Gayral P, Iskra-Caruana ML (2009) Phylogeny of banana streak virus reveals recent and repetitive endogenization in the genome of its banana host (Musa sp.). J Mol Evol 69:65–80. https://doi.org/10.1007/s00239-009-9253-2
Geering AD, Mcmichael LA, Dietzgen RG, Thomas JE (2000) Genetic diversity among banana streak virus isolates from Australia. Phytopathology 90:921–927. https://doi.org/10.1094/PHYTO.2000.90.8.921
Geering AD, Olszewski NE, Harper G, Lockhart BE, Hull R, Thomas JE (2005) Banana contains a diverse array of endogenous badnaviruses. J Gen Virol 86:511–520. https://doi.org/10.1099/vir.0.80261-0
Geering AD, Parry JN, Thomas JE (2011) Complete genome sequence of a novel badnavirus, banana streak IM virus. Arch Virol 156:733–737. https://doi.org/10.1007/s00705-011-0946-7
Geijskes RJ, Braithwaite KS, Dale JL, Harding RM, Smith GR (2003) Sequence analysis of an Australian isolate of sugarcane bacilliform badnavirus. Arch Virol 147:2393–2404. https://doi.org/10.1007/s00705-002-0879-2
Harper G, Hull R (1998) Cloning and sequence analysis of banana streak virus DNA. Virus Genes 17:271–278. https://doi.org/10.1023/A:1008021921849
Harper G, Osuji JO, Heslop-Harrison JS, Hull R (1999) Integration of banana streak badnavirus into the Musa genome: molecular and cytogenetic evidence. Virology 255:207–213. https://doi.org/10.1006/viro.1998.9581
Harper G, Hart D, Moult S, Hull R (2002) Detection of banana streak virus in field samples of bananas from Uganda. Ann Appl Biol 141:247–257. https://doi.org/10.1111/j.1744-7348.2002.tb00216.x
Harper G, Hart D, Moult S, Hull R, Geering A, Thomas J (2005) The diversity of banana streak virus isolates in Uganda. Arch Virol 150:2407–2420. https://doi.org/10.1007/s00705-005-0610-1
He Y, Ruan X, Zhao Q, Chen X, Liu F, Li H (2007) Analysis of genetic diversity of banana streak virus. In: proceeding of the annual meetng of Chinese society for plant pathology (2007). Yangling, Shanxi, pp 270–271
Hohn T, Fütterer J, Hull DR (1997) The proteins and functions of plant pararetroviruses: knowns and unknowns. Crit Rev Plant Sci 16:133–161. https://doi.org/10.1080/07352689709701947
Iskra-Caruana ML, Chabannes M, Duroy PO, Muller E (2014) A possible scenario for the evolution of banana streak virus in banana. Virus Res 186:155–162. https://doi.org/10.1016/j.virusres.2014.01.005
James AP, Geijskes RJ, Dale JL, Harding RM (2011) Molecular characterisation of six badnavirus species associated with leaf streak disease of banana in East Africa. Ann Appl Biol 158:346–353. https://doi.org/10.1111/j.1744-7348.2011.00466.x
Jaufeerally-Fakim Y, Khorugdharry A, Harper G (2006) Genetic variants of banana streak virus in Mauritius. Virus Res 115:91–98. https://doi.org/10.1016/j.virusres.2005.06.015
King AMQ, Adams MJ, Lefkowitz EJ, Carstens EB (2012) Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Elsevier/Academic Press
Kubiriba J, Legg JP, Tushemereirwe W, Adipala E (2001) Vector transmission of banana streak virus in the screenhouse in Uganda. Ann Appl Biol 139:37–43. https://doi.org/10.1111/j.1744-7348.2001.tb00128.x
Lockhart BE (1995) Banana streak badnavirus infection in Musa: epidemiology, diagnosis, and control. Food Fert Tech Bull 143
Lockhart BEL, Autrey LJC (1988) Occurrence in sugarcane of a bacilliform virus related serologically to banana streak virus. Plant Dis 72:230–233. https://doi.org/10.1094/PD-72-0230
Lockhart BEL, Autrey LJC, Comstock JC (1992) Partial purification and serology of sugarcane mild mosaic virus, a mealybug-transmitted closterolike virus. Phytopathology 82:691–695. https://doi.org/10.1094/Phyto-82-691
Lyttle DJ, Orlovich DA, Guy PL (2011) Detection and analysis of endogenous badnaviruses in the New Zealand flora. Aob Plants 2011:567–588. https://doi.org/10.1093/aobpla/plr008
Muller E, Dupuy V, Blondin L, Bauffe F, Daugrois J, Nathalie L, Iskra-Caruana ML (2011) High molecular variability of sugarcane bacilliform viruses in Guadeloupe implying the existence of at least three new species. Virus Res 160:414–419. https://doi.org/10.1016/j.virusres.2011.06.016
Ndowora T, Dahal G, Lafleur D, Harper G, Hull R, Olszewski NE, Lockhart B (1999) Evidence that badnavirus infection in Musa can originate from integrated pararetroviral sequences. Virology 255:214–220. https://doi.org/10.1006/viro.1998.9582
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mole Biol Evol 3:418–426
Ortiz R (1996) The potential of AMMI analysis for field assessment of Musa genotypes to virus infection. Hortscience 31:829–832
Rothnie HM, Chapdelaine Y, Hohn T (1994) Pararetroviruses and retroviruses: a comparative review of viral structure and gene expression strategies. Adv Virus Res 44:1–67. https://doi.org/10.1016/S0065-3527(08)60327-9
Sharma SK, Kumar PV, Poswal R, Rai R, Geetanjali AS, Prabha K, Jain RK, Baranwal VK (2014) Occurrence and distribution of banana streak disease and standardization of a reliable detection procedure for routine indexing of banana streak viruses in India. Sci Hortic 179:277–283. https://doi.org/10.1016/j.scienta.2014.09.043
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Xie L, He Y, Chen X, Zhao Q, Ruan X, Li H (2007) Analysis of genomic sequence and characteristics of banana streak virus from Guangdong. In: proceeding of the annual meeting of Chinese Society for Plant Pathology (2007), Yangling, Shanxi. pp. 267–269
Zhuang J, Wang J, Zhang X, Liu Z (2011) Molecular characterization of banana streak virus isolate from Musa Acuminata in China. Virol Sin 26:393–402. https://doi.org/10.1007/s12250-011-3212-y
Funding
The study was supported part by Special Fund of China for Agro-scientific Research in the Public Interest (Grant No. 201203076–07) and National Key R&D Program of China (Grant No.2019YFD1001800).
Author information
Authors and Affiliations
Contributions
LHP designed the study and revised the manuscript. RXQ analyzed the data and wrote the draft of the manuscript. WZL analyzed the data. ZL, WW and SJ performed the experiments. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors. It is original and has not been published elsewhere.
Conflict of interest
All authors declare that no competing interests exist.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Fig. S1
Unrooted tree indicating the species identified in Fig. 1 grouped into two groups according to the criterion of nucleotide sequence identities of greater than 72% based on the BSV RT/RNase H region. Details of the viruses are provided in Table S1. CYMV (citrus yellow mosaic badnavirus), CSSV (cacao swollen shoot virus), DBV (dioscorea bacilliform virus), ComYMV (commelina yellow mottle virus), SCBV (sugarcane bacilliform virus), and TaBV (taro bacilliform virus) are used as the outgroups. (PNG 3639 kb)
Fig. S2
The phylogeny tree was established with the nucleotide sequences (A) and amino acid (B) sequences of ORF 2 of DN5–1 and DN5–4 isolates from China. The trees were constructed using the neighbor-joining method. The numbers at the nodes indicate bootstrap support (1000 replicates). Details of the viruses are given in Table S1. KTSV (kalanchoe top-spotting virus), ComYMV (commelina yellow mottle virus), TaBV (taro bacilliform virus), BCVBV (bougainvillea chlorotic vein banding virus), DBSNV (dracaena bacilliform SN virus), CYMV (citrus yellow mosaic virus), CSSV (cacao swollen shoot virus), and RTBV (rice tungro bacilliform virus) are used as the outgroups. (PNG 223 kb)
Fig. S3
Phylogenetic analysis of the nucleotide sequences (A) and amino acid (B) sequences of ORF 3 of DN5–1 and DN5–4 isolates from China. The trees were constructed using the neighbor-joining method. The numbers at the nodes indicate bootstrap support (1000 replicates). Details of the viruses are given in Table S1. KTSV (kalanchoe top-spotting virus), ComYMV (commelina yellow mottle virus), TaBV (taro bacilliform virus), BCVBV (bougainvillea chlorotic vein banding virus), DBSNV (dracaena bacilliform SN virus), CYMV (citrus yellow mosaic virus), CSSV (cacao swollen shoot virus), and RTBV (rice tungro bacilliform virus) are used as the outgroups. (PNG 259 kb)
Table S1
Badnaviruses obtained from GenBank. (DOC 66 kb)
Rights and permissions
About this article
Cite this article
Rao, XQ., Wu, ZL., Wang, W. et al. Genetic diversity analysis reveals new badnaviruses infecting banana in South China. J Plant Pathol 102, 1065–1075 (2020). https://doi.org/10.1007/s42161-020-00646-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42161-020-00646-8