Abstract
The increasing reliance on electronic health record (EHR) in areas such as medical research should be addressed by using ample safeguards for patient privacy. These records often tend to be big data, and given that a significant portion is stored as free (unstructured) text, we decided to examine relevant work on automated free text de-identification with recurrent neural network (RNN) and conditional random field (CRF) approaches. Both methods involve machine learning and are widely used for the removal of protected health information (PHI) from free text. The outcome of our survey work produced several informative findings. Firstly, RNN models, particularly long short-term memory (LSTM) algorithms, generally outperformed CRF models and also other systems, namely rule-based algorithms. Secondly, hybrid or ensemble systems containing joint LSTM-CRF models showed no advantage over individual LSTM and CRF models. Thirdly, overfitting may be an issue when customized de-identification datasets are used during model training. Finally, statistical validation of performance scores and diversity during experimentation were largely ignored. In our comprehensive survey, we also identify major research gaps that should be considered for future work.
Similar content being viewed by others
Introduction
As the use and volume of medical records continues to rapidly grow in various areas, including research, there is a growing need to safeguard patient privacy for ethical and legal reasons [1]. In the USA, the confidentiality of patient information is legislated by the Health Insurance Portability and Accountability Act (HIPAA) [2]. The act lists 18 categories of protected health information (PHI), such as telephone numbers, geographic data, social security numbers, email addresses, and full face photos [3], that require special attention (see Table 1). PHI is health information capable of being linked, through the operations of a HIPAA-covered entity or business associate of the entity, to an individual patient.
In the HIPAA world, the de-identification of PHI involves the reduction of risk to an acceptable level not subject to predefined privacy restrictions [4]. This process is carried out through the Expert Determination Method or the Safe Harbor method [5]. The Expert Determination method requires the opinion of a qualified statistician to determine whether the risk of re-identification is very small. Our survey paper is concerned with the Safe Harbor method, which aims to remove specific identifiers (18 categories that make up PHI) from medical data. De-identification and scrubbing [6] are synonymous terms used in the context of medical data research. Within the same scope, anonymization and de-identification can be considered synonymous, but there is a subtle difference between the two terms [7]. The former is not reversible, whereas de-identification is reversible. Only skilled personnel should perform manual de-identification, a process that is tedious, error-prone, and expensive [1]. For the removal of PHI from a large corpus of records, automated de-identification is a better alternative.
With regard to PHI in the form of text, this information can be represented as structured data (lab results, patient demographic data, etc.), free text (emails to schedule appointments, symptoms as described by patients, etc.), or a combination of both, and is stored in electronic health record (EHRs) [8]. The healthcare industry is experiencing a growth of big data EHRs [9], with a significant portion being stored as free text [10]. Some people may mistakenly believe that 1,000,000 records of structured data, which is comfortably in the big data category [11], is comparable in size to 1,000,000 records of free text. Rather than simply stating this notion is false, we provide an appreciable example; a single record of unstructured data for an individual’s genome contains about 1 terabyte of information [12]. A trove of published papers exists on the automated removal of PHI from structured data. However, the international research focus seems to be currently transitioning from structured data to unstructured data, which is a more challenging task as there are no limitations to the format of free text.
The automated de-identification of free text can follow a rule-based approach, machine learning approach, or a combination of both [13]. In general, rule-based approaches incorporate pattern matching, regular expressions, and dictionary lookups [10]. Rule-based methods require little or no labeled (annotated) data but usually have limited generalizability. Machine-learning methods, by contrast, have the advantage of wider generalizability but often involve supervised learning and a large corpus of training data.
To present an up-to-date survey on automated methods for the de-identification of free text, we exhaustively examined peer-reviewed research papers that date back five years (January 2015–June 2020). De-identification methods embracing machine learning approaches and hybrid machine learning/rule-based approaches were considered, while pure rule-based approaches were ignored. We observed that works utilizing recurrent neural network (RNNs) and conditional random field (CRFs) comprised an overwhelming majority, thus indicating the popularity of these machine learning methods. The further narrowing of curated papers to focus exclusively on RNNs and CRFs increases the novelty value of this survey.
For the most part, RNN models, especially long short-term memory (LSTM) networks, outperformed CRF and rule-based models. We note that the evidence is inconclusive about any advantage gained from using hybrid or ensemble models that incorporate LSTM-CRF systems. In two studies with comparatively high overall F-measure scores, overfitting [14] may have occurred, possibly arising from the use of a customized de-identification dataset. Further studies are needed to investigate this issue. Unfortunately, for performance scores across the board the use of statistical validation has been practically ignored, which is a cause for concern as the determination of statistical significance adds clarity. In addition, many researchers did not appear to be sufficiently invested in diversifying the approach to their respective works. For example, experimenting with only 4 categories of PHI out of 18 possible categories, as defined by HIPAA, may not be comparatively useful.
The remainder of this paper is organized as follows: “Recurrent neural networks: brief introduction” section provides a brief introduction to RNN networks; ’Conditional random fields: Brief introduction” section gives a brief introduction to CRF networks; “Recurrent neural networks: methods for medical free text de-Identification” section describes and analyzes RNN approaches for the automated de-identification of medical free text; “Conditional random fields: methods for medical free text de-Identification section describes and analyzes CRF approaches for the automated de-identification of medical free text; “Discussion of surveyed works” section discusses findings of our survey work, identifies gaps in the current research, and explains the performance metrics used in the curated works; and “Conclusion” concludes with the main points of the paper and offers suggestions for future work.
Recurrent neural networks: brief introduction
Artificial neural network (ANNs) are nonlinear models inspired by the biological neural structure of the brain. The ANN consists of interconnected nodes and is a directed graph where each node i initiates a transfer function \(f_i\) (sigmoid, Heaviside, etc.) to produce an output \(y_i\) [15]. There is a connection weight between the nodes and also a threshold bias. The feedforward ANN is a simple neural network, with information moving only in the forward direction from the input, through hidden nodes (usually present), and then to the output. The relative ease with which ANNs can project a predefined vocabulary into hidden nodes in order to semantically associate similar words is noteworthy [16].
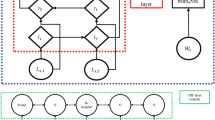
While feedforward networks are unable to maintain state, RNNs can use internal state to process input sequences [17]. Unlike feedforward networks, RNNs have cyclic or feedback connections that facilitate updates to their current state based on previous states and current inputs. This makes RNNs more efficient in sequence modeling tasks. For natural language processing (NLP) operations, an advantage of using RNNs is their ability to remember words presented during an earlier iteration, a helpful feature when determining context [16]. Figure 1, originally illustrated by Mulder et al. [16], clearly shows the difference between an RNN network and a feedforward network.
RNN (left) vs. feedforward (right) [16]
RNNs, however, suffer from the exploding and vanishing gradient problems, which limits modeling of long-term dependencies to no more than 10 discrete time steps between input signals and their output [18]. To get around this obstacle, a specialized RNN model, the LSTM, was introduced. The original LSTMs contained memory blocks in the recurrent hidden layer, with each block housing an input gate and output gate. Later models were reinforced by the addition of forget gates and peephole connections.
Promising results with RNNs now mostly originate from LSTM networks [19]. An increasing number of classification and prediction tasks that assimilate time series data use LSTM models. These tasks include speech recognition, sentence embedding, and correlation analysis. Bidirectional LSTMs, which feed each training sequence backward and forward to two separate recurrent nets, are especially popular [20,21,22].
Conditional random fields: brief introduction
Many models selected for NLP tasks must be able to predict multiple interdependent variables [23]. In part-of-speech tagging [24], for example, each variable \(y_p\) of an output vector y is the tag at position p of the word. The observed feature x is split into input attributes, each denoted by \(x_p\), and contains information about the specific word, such as its identity, prefixes and suffixes.
Graphical models are a convenient way to demonstrate the connectivity between output variables [25]. A lot of research work in this area is centered on generative models, such as the hidden Markov model (HMM) [26], which characterize a joint probability distribution over observed input features x and corresponding annotated outputs y. Effectively modeling this distribution means that all possible values of x must be used, an often intractable operation due to the high dimensionality of x. Moreover, the nature of the dependencies between features may become excessively entangled and hinder development of a useful model.
The issues highlighted in the previous paragraph are addressed through direct modeling of the conditional distribution p(y|x) [23]. Classifiers such as logistic regression [27] are also discriminative since they benefit from conditional distribution. CRFs can therefore be described as a framework for building probabilistic models that ignore interdependencies between input values [28].
CRFs and ANNs [15] both fit into the category of discriminative, probabilistic models and are relatively similar in that regard. Although ANNs have seen an explosive rise in popularity as classifiers, these networks are also capable of predicting multiple outputs [23]. ANNs and CRFs can be trained via the same methods, but ANNs use a shared hidden representation [29] to model the dependence between output variables, whereas CRFs rely on direct functions of output variables to learn this relationship. In other words, a CRF does not require any hidden layers for efficient training. CRFs are used for various tasks such as image denoising [30], phishing detection [31], and musical audio-to-score alignment [32]. In Figs. 2 and 3, two different configurations of linear-chain CRFs, originally illustrated by Sutton and McCallum [23], are shown. Transitional states based on current observations are shown in Fig. 2, while transitional states based on all observations are shown in Fig. 3.
CRF-transitional states depend on current observation [23]
CRF-transitional states depend on all observations [23]
Recurrent neural networks: methods for medical free text de-identification
In this section, we describe various RNN strategies for the de-identification of medical free text. Works of research are presented in alphabetical order by author. All scores obtained from metrics (F-measure, etc.) are implicitly reported as percentages, and the highest or best scores that we discuss are associated with overall PHI categories rather than individual PHI categories. If the authors of a research paper have proposed an RNN model that outperforms a CRF model, the work is included only in this section. If the proposal integrates both RNN and CRF algorithms into one model, the work is included both in “Conditional random fields: methods for medical free text fe-Identification” section, which discusses de-identification of medical free text with CRFs, and this section. Lastly, if the proposal is a CRF model that outperforms an RNN model, the work is included only in “Conditional random fields: methods for medical free text fe-Identification” section.
Table 2 provides an alphabetical listing by author of the papers discussed in this section. Comparisons between performance scores for separate works of research or separate experiments within the same paper may not be valid. However, providing these scores may be valuable for future comparative research.
Dernoncourt et al. 2017 [33] (De-identification of patient notes with recurrent neural networks)
Three models (bidirectional LSTM, CRF, combined bidirectional LSTM-CRF) were evaluated on the 2014 i2b2 [34] de-identification dataset and a customized version of the MIMIC de-identification dataset [35]. The popular 2014 i2b2 dataset was created for the 2014 i2b2/UTHealth shared task challenge. It is a corpus of 1,304 longitudinal clinical records (790 for training, 514 for testing) and about 28,900 PHI instances. Table 3 provides an overview of this de facto benchmark dataset. The customized MIMIC dataset contains 1,635 discharge summaries, each summary associated with a different patient, and about 60,700 PHI instances. Training set sizes were purposely varied, with analyses done on the i2b2 dataset for six PHI categories and on the MIMIC dataset for five PHI categories. With regard to the LSTM model, stochastic gradient descent [36], and hyper-parameter tuning were involved in the training method. Hand-crafted features [37], similar to those of the top performers in the i2b2 challenge, were used with the CRF model during training. The LSTM model outperformed the combined model, which in turn outperformed the CRF model. For LSTM, the highest F-measure score (99.23) was obtained with a precision of 99.21 and a recall of 99.25. These high scores are based on the customized MIMIC dataset, and further experimentation may be required to determine if any overfitting occurred. In comparison, the highest F-measure score obtained with the 2014 i2b2 dataset was 97.88.
Jiang et al. 2017 [38] (De-identification of medical records using conditional random fields and long short-term memory networks)
Two de-identification systems (CRF, LSTM) used in the 2016 CEGS N-GRID Shared Tasks Track 1 challenge [49] were evaluated in this work. For the CRF system, manually extracted features were involved in training. The LSTM system, which was bidirectional, used classifying tags for each represented token. Stochastic gradient descent and hyper-parameter tuning were involved in the training of the LSTM system. Both systems were evaluated on the CEGS N-GRID 2016 Shared Task 1 dataset, with the raw text being pre-processed before being fed to the models in order to remove seven categories of PHI. The CEGS N-GRID 2016 dataset is a corpus of 1000 psychiatric intake records (600 for training, 400 for testing) and about 34,400 PHI instances. Table 4 gives an overview of this de facto benchmark dataset. According to the results, the pre-processing step improved the performance of both systems. The results also show that the LSTM model convincingly outperformed the CRF model. The highest F-measure score obtained for LSTM was 91.45, with accompanying precision and recall scores of 93.24 and 89.72, respectively. The researchers have therefore concluded that their LSTM system is the better model. Going by their paper alone, this conclusion should not be generalized. At the very least, the researchers should provide evidence indicating that the best CRF model in the challenge has also underperformed their LSTM model.
Kajiyama et al. 2018 [39] (De-identifying free text of Japanese electronic health records)
Using three datasets, the researchers examined the removal of five PHI categories through a rule-based approach, a CRF approach, and a bidirectional LSTM approach. The first dataset was sourced from the NTCIR-10 MedNLP De-identification Task [50], the second was a synthetic dataset annotated by the researchers, and the third was a combination of the two datasets. The combined dataset, which contained 82 records, was used for training. No information was provided on the number of PHI instances for this dataset. The researchers implemented their own rule-based approach, and for the CRF implementation, the mallet libraryFootnote 1 was used. In the LSTM approach, the neural network was trained with feature vectors, and the output was subsequently processed by a CRF model using character level tags. Thus, it should be noted that this model is essentially a hybrid as it integrates both LSTM and CRF algorithms. Based on the consistency of results, the researchers selected the hybrid model (F-measure score of 80.61) over the rule-based and CRF alone models. However, the highest F-measure score in this research was actually obtained by the rule-based method. Precision and recall scores have not been provided in this paper, an omission that weakens the value of the work as these scores may provide additional insight.
Kim et al. 2018 [40] (Ensemble-based methods to improve de-identification of electronic health record narratives)
Three ensemble methods that combine machine learning approaches and rule-based approaches were used to de-identify seven PHI categories. The ensemble methods, each composed of 12 individual models, are voting [51], decision template [52], and stacked ensemble [53]. Pre-processing was carried out with the Stanford CoreNLP tool [54]. Individual machine learning models were trained on the 2014 i2b2 challenge dataset and include CRF, LSTM, LSTM-CRF, support vector machine (SVM) [55], and margin improved relaxed algorithm (MIRA) [56]. The stacked ensemble recorded the highest F-measure score (95.73), along with precision and recall scores of 97.04 and 94.45, respectively. This groundbreaking study could benefit from database diversity. Subsequent de-identification evaluations of the stacked ensemble method should consider multiple datasets from different clinical sources.
Lee et al. 2016 [41] (Feature-augmented neural networks for patient note de-identification)
This work stands on the preprint version of the research later published by Dernoncourt et al. [33], which we discussed earlier in the section. Lee at al. made their LSTM model more robust by adding human-engineered features, along with standard features from an EHR database. The model was evaluated on the customized version of the MIMIC dataset originally presented by Dernoncourt et al. and trained using stochastic gradient descent and hyper-parameter tuning. For the de-identification of 12 PHI categories, Lee et al. experimented on a base model of no features, one model of only EHR features, and another of both EHR and hand-crafted features. The researchers discovered that the addition of features improved performance. The highest F-measure score (99.26) came from the model with both EHR and hand-crafted features and corresponded to a precision of 99.21 and a recall of 99.31. Just like the work of Dernoncourt et al., the scores in this study seem very high, and more experimentation may be necessary to establish if overfitting has occurred.
Lee et al. 2019 [42] (An empirical test of GRUs and deep contextualized word representations on de-identification)
Gated recurrent unit (GRUs) [57] and deep contextualized word representations [58] are introduced by the researchers as an alternative to bidirectional LSTMs for de-identification. The main advantage that GRUs, which are also RNNs, have over LSTMs is a simpler architecture. Whereas an LSTM structure may consist of forget, update, and output gates, a GRU structure may only contain reset and update gates [42]. Deep contextualized word representations can provide token embeddings with context. Performance was evaluated on the 2016 CEGS N-GRID dataset and the 2014 i2b2 dataset, both being divided into training and test sets to assess the de-identification of 5 PHI categories. The researchers found that the substitution of GRUs for LSTMs has no significant effect on performance. The highest F-measure score (89) was recorded for the LSTM unit, in conjunction with a precision of 90.88 and a recall of 87.20. In terms of PHI diversity, the availability of only 5 categories for de-identification slightly diminishes the impact of this study.
Liu et al. 2017 [43] (De-identification of clinical notes via recurrent neural network and conditional random field)
In the 2016 CEGS N-GRID Shared Tasks Track 1 challenge, the proposal by Liu et al. secured first place. The model also performed notably in the 2014 i2b2 challenge. Their method is a combination of three machine learning subsystems (bidirectional LSTM without hand-crafted features [21], bidirectional LSTM with hand-crafted features [59], CRF with hand-crafted features) along with a rule-based system. The hybrid system was tasked with removing seven categories of PHI from the 2016 CEGS N-GRID Shared Tasks Track 1 dataset. Stochastic gradient descent and hyper-parameter tuning were used in the training of the LSTM models. Individually, both LSTM subsystems outperformed the CRF subsystem, with the LSTM containing hand-crafted features being the top performer. The highest F-measure score (96.98) obtained by the hybrid system corresponded to a precision of 97.94 and a recall of 96.04. It would be interesting to compare the performance of this award-winning entry with other entries from the 2016 N-GRID challenge, if the task involved the removal of more than seven categories of PHI.
Madan et al. 2018 [44] (Redaction of protected health information in EHRs using CRFs and Bi-directional LSTMs)
Evaluations on both CRF and bidirectional LSTM models with the 2014 i2b2 dataset were done to determine which was better at removing four PHI categories. Both systems had a pre-processing stage consisting of sentence detection and tokenization. The CRF model was trained using the limited memory Broyden-Fletcher-Goldfarb-Shanno (BFGS) algorithm [60] and required hand-crafted features. These features were not needed for the LSTM model. With an F-measure score of 95.92, the LSTM approach outperformed the CRF approach. Precision and recall scores were not provided for the overall PHI categories, which is a shortcoming of this paper. Furthermore, the low number (four) of PHI categories presented for de-identification impacts the validity of this research.
Richter et al. 2019 [45] (Deep learning approaches outperform conventional strategies in de-identification of German medical reports)
Adopting a rule-based method with trained statistical capabilities as a baseline [61], the researchers used a cardiology dataset of 113 records and about 5,200 PHI instances to compare the performance of a bidirectional LSTM model with a CRF model. About 75% of the records were used for training data and the remainder for test data. The CRF algorithm was accessed via the sklearn-cfsuite wrapper Footnote 2. To implement the LSTM algorithm, Keras Footnote 3 and Tensorflow Footnote 4 were used. Training involved hyper-parameter tuning. For the eight PHI categories de-identified, the LSTM model had the highest F-measure score (96.0), along with a precision of 97.0 and a recall of 95.5. This study could have been strengthened with the use or addition of a gold-standard dataset, such as the 2014 i2b2.
Shweta et al. 2016 [46] (A recurrent neural network architecture for de-identifying clinical records)
For the de-identification of seven categories of PHI from the 2014 i2b2 dataset, two variants of the RNN model, Elman-type [62] and Jordan-type [63], were compared to a baseline CRF model. In an Elman-type network, each state contains information about previous hidden layer states. In a Jordan-type network, inputs to recurrent connections are obtained from output posterior probabilities. Training of the RNN models involved stochastic gradient descent and hyper-parameter tuning. The CRF model was trained with a standard set of hand-crafted features. Both RNN variants outperformed the CRF model, with the Jordan-type being the top performer: precision (97.26), recall (90.67), and F-measure (93.84). Since both variants have previously been shown to perform similarly [64], one should have been replaced with a dissimilar RNN variant to reinforce the credibility of this research.
Trienes et al. 2020 [47] (Comparing rule-based, feature-based and deep neural methods for de-identification of Dutch medical records)
Upon constructing a dataset of sampled records from nine Dutch healthcare institutions, the researchers investigated the de-identification of eight PHI categories through a rule-based approach, a CRF approach, and a bidirectional LSTM approach. Twelve experts annotated the dataset, which contained 1260 records and about 17,500 PHI instances. To evaluate performance between English and Dutch datasets, the nursing notes corpus dataset [1] (2,434 records, about 1,800 PHI instances) and the 2014 i2b2 dataset were also used. DEDUCE, a rule-based approach developed for Dutch medical records, was adopted by the researchers [65]. For the CRF approach, a subset of features from a token-based approach by Liu et al. [37] was utilized. In the LSTM approach, training of the model involved stochastic gradient descent and hyper-parameter tuning. Since this method was also merged with CRF architecture, the bidirectional LSTM model should rightly be referred to as a hybrid. For all three datasets, the hybrid model outperformed the rule-based method and plain CRF method. The highest F-measure score (91.20) was derived from a precision of 95.90 and a recall of 86.90. We note that one of the PHI categories in this study is called “other.” As the “other” category has not been clearly defined, this presents a research comparison problem in an otherwise informative paper.
Yang et al. 2019 [48] (A study of deep learning methods for de-identification of clinical notes in cross-institute settings)
Using a bidirectional LSTM-CRF model to de-identify 10 PHI categories, the researchers compared 5 word embeddings and 2 customized approaches. Word embedding maps words to vectors of real numbers and can capture features in a low-dimensional matrix, thus eliminating or reducing the need for feature engineering. The training set was obtained from the 2014 i2b2 challenge, while the test set came from the University of Florida (UF) Health Integrated Data Repository Footnote 5. Word embeddings were sourced from GoogleNews [66], CommonCrawl [67, 68], MIMIC-word2vec [69], MIMIC-fastText [67, 69], and MADE [70]. The first customized approach entailed combining the UF and i2b2 datasets, and the second involved fine-tuning the model based on the i2b2 dataset with UF data. Stochastic gradient descent and hyper-parameter tuning were used to train the hybrid model. Fine-tuning of the 12b2 model with UF data emerged as the better of 2 approaches, while CommonCrawl, a general English word embedding, outperformed the other 4 embeddings. The highest F-measure score (96.46) for CommonCrawl corresponded to precision and recall scores of 97.97 and 94.98, respectively. Undoubtedly, this is exciting research, but the practice of customizing training approaches with data from the test source may limit the generalizability of this study.
Conditional random fields: methods for medical free text de-identification
In this section, we describe various CRF strategies for the de-identification of medical free text. Works of research are presented in alphabetical order by author. All scores obtained from metrics (F-measure, etc.) are implicitly reported as percentages, and the highest or best scores that we discuss are associated with overall PHI categories rather than individual PHI categories. If the authors of a research paper have proposed a CRF model that outperforms an RNN model, the work is included only in this section. If the proposal integrates both RNN and CRF algorithms into one model, the work is included both in “Recurrent neural networks: methods for medical free text de-Identification” section, which discusses de-identification of medical free text with RNNs, and this section. Lastly, if the proposal is an RNN model that outperforms a CRF model, the work is included only in “Recurrent neural networks: methods for medical free text de-Identification” section.
Table 5 provides an alphabetical listing by author of the papers discussed in this section. Comparisons between performance scores for separate works of research or separate experiments within the same paper may not be valid. However, providing these scores may be valuable for future comparative research.
Berg and Dalianis 2019 [71] (Augmenting a de-identification system for Swedish clinical text using open resources and deep learning)
Swedish researchers used three de-identification datasets to compare the performance of a CRF model to a bidirectional LSTM model. The datasets are listed as the Stockholm electronic patient records (EPR) PHI Corpus [78] (100 patient records, about 4400 PHI instances), Stockholm EPR PHI Domain Corpus [78] (number of records not provided, about 2900 PHI instances), and Stockholm Umea Corpus 3.0 [79] (102 records, about 31,000 PHI instances). For each set, training was performed on 90% of the records with testing done on the rest. Among the eight PHI categories the researchers intended to remove were “first name”, “last name”, “full date”, and “part date.” The CRF algorithm was implemented with CRFSuite Footnote 6 and the sklearn-cfsuite wrapper. Gradient descent with limited memory BFGS optimized the CRF model during training, and hyper-parameter tuning of the LSTM model occurred during training. CRF had the higher F-measure best score (91.00) of the two models, together with a precision of 94.66 and recall of 86.72. In this study, “first name” and “last name” counted as two separate PHI categories, and the same can be said for “full date” and “part date.” As per HIPAA, however, there is only a “names” category and a “dates” category. We point out that 10-fold cross validation was used for the CRF model, but the LSTM model was only evaluated on the first 3 folds because of time limitations. The disparity somewhat undermines the relative credibility of this work.
Bui et al. 2017 [72] (The UAB informatics institute and 2016 CEGS N-GRID de-identification shared task challenge)
After submitting a proposal to the 2016 CEGS N-GRID Shared Tasks Track 1 challenge, the researchers published a paper on the performance of their model, which secured fourth place. Using a hybrid approach that incorporates a CRF and rule-based system, the model relied on 4 main processing steps to de-identify 12 categories of PHI from the 2016 CEGS N-GRID dataset. The first step involves pre-processing, the second requires pattern-matching with regular expressions, the third relies on dictionary-matching, and the final uses the Stanford Named Entity Recognizer [80] implementation of CRF. Named entity recognition aims to locate and classify named entities into pre-existing categories. The highest score for F-measure (93.66) was derived from a precision of 96.29 and a recall of 91.18. By virtue of its fourth place rank, this approach is certainly notable. However, the research contribution of this paper would be more meaningful if information on how to improve the model, or perhaps how to beat the first-place model, was provided.
Bui et al. 2018 [73] (Is Multiclass automatic text de-identification worth the effort?)
A binary model was compared with two multi-class models to evaluate the efficiency of removal for seven PHI categories. The investigation was performed on the 2014 i2b2 de-identification dataset. The pre-processing stage involved tokenization and token class assignment. During the training stage, the Stanford Named Entity Recognizer was used to implement CRF. After processing, document readability was manually assessed by reviewers, with a pooled Kappa [81] implemented to estimate the inter-rater agreement. In terms of readability, the multi-class model showed no advantage over the binary model. The highest F-measure score (95.10) was attributed to the binary model. A precision of 98.50 and recall of 92.00 matched this F-measure score. The topic of this paper may be a bit misleading in that it appears to suggest a general solution. In actuality, the findings of this research only relate to CRF.
Du et al. 2018 [74] (A machine learning based approach to identify protected health information in Chinese clinical text)
Randomly selected discharge summaries from regional health centers in a Chinese province were used as training and test data to investigate the de-identification of seven PHI categories. The 2014 i2b2 Shared Tasks Track 1 provided corpus annotation guidelines on training and evaluating tagged entities. Two native Chinese speakers cross-annotated 14,719 records of discharge summaries to catch any missed PHI. The dataset was split into 11,775 records for training and 2,944 for testing, with the test containing about 25,400 PHI instances. No information was provided on the total number of PHI instances in the training set. As there is a post-processing rule-based stage after the CRF stage, the proposed model should be appropriately called a hybrid. The hybrid model, with scores of 99.27 (precision), 98.29 (recall), and 98.78 (F-measure), outperformed a plain CRF model. Ensuring that the training and test data came from different sources, instead of the same collection of discharge summaries, could enhance the robustness of this research.
Kajiyama et al. 2018 [39] (De-identifying free text of Japanese electronic health records)
See Kajiyama et al. 2018 in “Recurrent neural networks: methods for medical free text de-Identification” section.
Kim et al. 2018 [40] (Ensemble-based methods to improve de-identification of electronic health record narratives)
See Kim et al. 2018 in “Recurrent neural networks: methods for medical free text de-Identification” section.
Lee et al. 2017 [75] (A hybrid approach to automatic de-identification of psychiatric notes)
This work describes the combined CRF and rule-based system submitted to the 2016 CEGS N-GRID Shared Tasks Track 1 challenge for the removal of seven PHI categories. A pre-existing baseline system used one CRF tagger, while the hybrid system used two CRF taggers and one rule-based tagger. Against other participants in the challenge, the hybrid system placed second, achieving an F-measure score of 90.74, with precision and recall scores of 93.39 and 88.3, respectively. Although this paper is well-written, no information is provided about the scores of the winning entry, which could be significantly better or slightly better than Lee et al.’s model. No information is also given about the lower-placed entries.
Lee et al. 2017 [76] (Leveraging existing Corpora for de-identification of psychiatric notes using domain adaptation)
Domain adaptation [82] is used with a token-based CRF approach to reduce annotation costs for the training data in the target domain. Three different domain adaptation methods were used: instance weighting [83], instance pruning [83], and feature augmentation [84]. The de-identification task involved removing eight categories of PHI. Three different datasets made up the source domain: diabetes notes from the 2014 i2b2 De-identification Track 1 (470 out of 1304 original records, about 8900 PHI instances), discharge summaries from the 2006 i2b2 De-identification Track 1 challenge [85] (604 out of 889 original records, about 13,200 PHI instances), and outpatient notes from the University of Texas Health Science Center at Houston (325 records, about 10,500 PHI instances). The target domain (600 records, about 11,900 instances) came from a corpus of psychiatric notes provided by the 2016 CEGS N-GRID Shared Tasks Track 1 challenge. After pre-processing, the CRF model attempted to tag all instances of PHI. The feature augmentation method yielded the best performance: precision (93.46), recall (87.53), and F-measure (90.40). This is an engaging paper, and future work should investigate how the combined CRF-feature augmentation method compares with other CRF techniques that do not use domain adaptation.
Liu et al. 2017 [43] (De-identification of clinical notes via recurrent neural network and conditional random field)
See Liu et al. 2017 in “Recurrent neural networks: methods for medical free text de-Identification” section.
Phuong et al. 2016 [77] (A hybrid semi-supervised learning approach to identifying protected health information in electronic medical records)
A CRF model and a rule-based system were combined to analyze the de-identification of eight PHI categories based on the 2006 i2b2 dataset. Of the 889 records (about 19,500 PHI instances) in the dataset, 669 records were used for training and the remainder for testing. Six phases constituted this semi-supervised learning approach: (1) Supervised learning with CRFs and k-fold cross validation [86]; (2) Identification of PHI; (3) Rule-based processing; (4) Selection of records; (5) Updating of unlabeled records; and (6) Enhancing of labeled records. Compared to other approaches used in the experiment, the proposed hybrid solution had the highest recall score of 94.16. However, scores for precision (97.91) and F-measure (96.00) for this model were below those obtained from a plain CRF approach (98.93 and 96.02, respectively). In the absence of suitable statistical analysis, it cannot be determined with great confidence that the researchers’ recommendation is the best choice.
Trienes et al. 2020 [47] (Comparing rule-based, feature-based and deep neural methods for de-identification of Dutch medical records)
See Trienes et al. 2020 in “Recurrent neural networks: methods for medical free text de-Identification” section.
Yang et al. 2019 [48] (A study of deep learning methods for de-identification of clinical notes in cross-institute settings)
See Yang et al. 2019 in “Recurrent neural networks: methods for medical free text de-Identification” section.
Discussion of surveyed works
RNN models, particularly LSTM, have demonstrated their superiority over CRF and rule-based models in this survey. For the study by Berg and Dalianis [71] where CRF was shown to be better than LSTM, the researchers admitted that the LSTM model performs reasonably well. Furthermore, 10-fold cross validation was used for both of their models, but the LSTM network was only evaluated on the first 3 folds. In general, LSTMs appear to more adept than CRFs at recognizing semantic variance.
The combined use of LSTM and CRF systems in hybrid or ensemble models is an active area of research. Table 6, a common subset from Tables 2 and 5, lists papers where these combined models were the top performers, either among variations of themselves or in comparison to other models without LSTM-CRF systems. Although the work of Dernoncourt et al. [33] includes an LSTM-CRF hybrid, the paper does not feature in Table 5 (CRF only) or Table 6 (CRF-LSTM joint approach) because the hybrid model was not the top performer, and therein lies the problem. Within the scope of this survey, no evidence suggests that the combined models are the best performers in relation to other architectures that do not incorporate an LSTM-CRF system. According to Dernoncourt et al. [33], the plain LSTM approach works best. However, from a consistency perspective, Kajiyama et al. [39] recommend their hybrid method, but the highest F-measure score was recorded for their rule-based system. Trienes et al. [47] are the only researchers whose results favor a combined LSTM-CRF approach.
It is possible that the top LSTM models selected by Dernoncourt et al. [33] and Lee et al. [41] are overfitted. Overfitting occurs when a model memorizes data from a training set and cannot reliably generalize to unseen data. The highest F-measure scores for both models are above 99.2, the result of a train-test evaluation method on a customized MIMIC dataset. When Dernoncourt et al. [33] performed a train-test evaluation on the gold-standard 2014 i2b2 dataset, the highest F-measure score obtained was 97.88, an interesting point that supports our contention. As stated previously, more experimentation is required to determine if overfitting occurred.
Most noticeably, the highest overall score for F-measure dictates the choice of model in some papers, while model selection for other papers hinges on highest overall scores for both F-measure and recall. In some cases, the model selected through F-measure differs from the model selected via recall. This stems from the fact that the F-measure score associated with the highest recall score is lower (marginally lower in our surveyed papers) than the highest overall F-measure score. Concerning the de-identification of medical text, recall is viewed as more important than precision because PHI should never be exposed to unauthorized parties. Readers are reminded that the scores shown in Tables 2, 3, 4, 5, 6 are based on highest overall F-measure. The reporting of precision and recall throughout the compiled works was not consistent, but F-measure was always provided. Please see “Performance metrics” section for an explanation of precision, recall, and F-measure.
Among the surveyed works there is a lack of statistical analysis of the overall PHI scores for de-identification. Determining the statistical significance of these scores adds clarity, and there are some excellent methods, including Analysis of variance (ANOVA) [87] and Tukey’s Honestly Significant Difference (HSD) [88], that are popular within the research community. ANOVA tests whether the means of one or more independent factors are significant. Tukey’s HSD assigns group letters to means that are significantly different from each other.
As a minor point to note, there is a wide variation in the information provided by authors on the respective de-identification datasets that they use. We expected to encounter a moderate level of inconsistency since there is obviously no universal standard on reporting dataset information, but unfortunately, the issue borders on the extreme. On one hand, researchers such as Trienes et al. [47] provide adequate information, and on the other hand, researchers such as Kajiyama et al. [39] are not as forthcoming. The inconsistency makes the process of drawing comparisons between the surveyed works more challenging.
Last but not least, we emphasize the importance of diversity during experimentation. For instance, a study investigating the de-identification of 4 PHI categories lacks diversity. Putting our observation into context, readers should be reminded that HIPAA has defined 18 PHI categories. Hence, we recommend that at least 8 PHI categories should be considered. A more common example relates to the use of only one de-identification dataset in a study. We advise that at least two datasets should be involved.
Gaps in current research
There are significant gaps in the current research on de-identification of medical free text with RNN and CRF methods. The literature is lacking in research methods on topics such as big data, data streaming, data quality, and concept drift. We expound on those topics in the following paragraphs.
The increasing global dependence on big data signals that efficient and effective methods should exist to handle the many ways by which humans interact with this type of data [11]. It is important to stress that models constructed with regular data may not be useful for processing big data. For example, with reference to the de-identification of medical free text using LSTM, a model trained on a 10 kilobyte dataset of 1,000 EHRs may produce an overwhelming number of false negatives when processing a 10 terabyte dataset of 1,000 EHRs. Specific properties [89] define big data, such as volume, veracity, velocity, variety, variability, and value, and these should be factored into future research with big data in relation to some of the approaches discussed in this survey.
For all the surveyed works, the predictive models were trained on static data processed in an offline mode. However, online processing is necessary to handle streams of big data capable of overwhelming their respective computer systems. Significant research has been fueling the development of streaming methods and also Internet of Things (IoT) devices that allow for observations and comparisons in real time [90, 91]. Procedures such as incremental training or data batching are often used to train predictive models in these cases [92].
Data quality can be affected by insufficient, missing, erroneous, and redundant data, among other issues [93]. These concerns may complicate the process of model training and reduce the efficiency of trained models. Detection of data quality problems in EHRs of free text is a fertile area for exploration with LSTM and CRF methods.
Also known as dataset shift, concept drift refers to the temporal variation of data distributions [94]. For instance, an LSTM or CRF model trained today to de-identify medical free text may have a lower optimum recall score in 30 years. This could be due, perhaps, to a radically different format being used for a HIPAA PHI category, such as for vehicle identifiers or device identifiers. Research investigating the effect of time on such de-identification models is a very promising area. Figure 4, originally illustrated by Chilakapati [95], portrays concept drift as a divergence of model and data decision boundaries that leads to a loss of predictability.
Concept drift Illustration [95]
Performance metrics
F-measure scores are used in all the surveyed works, with precision and recall scores provided by most of the respective authors. In order to explain these three metrics, it is necessary to start with the fundamental metrics and then build on the basics. Our list of applicable performance metrics is explained as follows:
-
A True Positive (TP) is a positive instance correctly identified as positive.
-
A True Negative (TN) is a negative instance correctly identified as negative.
-
A False Positive (FP), also known as Type I error, is a negative instance incorrectly identified as positive.
-
A False Negative (FN), also known as Type II error, is a positive instance incorrectly identified as negative.
Based on these fundamental metrics, the other performance metrics are derived as follows:
-
Recall, also known as sensitivity, is equal to \(\hbox {TP} / (\hbox {TP} + \hbox {FN})\).
-
Precision, also known as positive predictive value, is equal to \(\hbox {TP} / (\hbox {TP} + \hbox {FP})\).
-
Traditional F-measure, also known as the harmonic mean of precision and recall, is equal to \(2 \cdot Precision \cdot Recall / (Precision + Recall)\).
-
The general formula for F-measure, where Recall is more significant that Precision by a positive real factor of \(\beta \), is equal to \((1+\beta ^2) \cdot Precision \cdot Recall / (\beta ^2 \cdot Precision + Recall)\).
Conclusion
The proliferation of EHRs in various areas such as medical research should be covered by adequate protections for patient privacy. As these medical records are often big data, an additional layer of complexity must be catered for during the de-identification of PHI. Since a considerable number of medical records are stored as free text, we decided to do a survey on automated free text de-identification. We discovered that the curated works utilized RNN and/or CRF, two machine learning methods. Our analysis led to several informative findings.
RNN models, particularly LSTM, generally perform better than CRF and rule-based models. When LSTM and CRF models were combined in various studies, the evidence was inconclusive about any advantage gained. We suspect that overfitting occurred in works where the overall F-measure score was uncommonly high. This may be due to the customization of the popular MIMIC dataset. Throughout the surveyed works, interestingly, scant attention was paid to the statistical analysis of PHI de-identification scores. In addition, not enough consideration was given to diversity during experimentation. Finally yet importantly, there is a lack of de-identification studies for medical free text pertaining to big data, data streaming, data quality, and concept drift, just to name a few topics, and future work should address these gaps.
Data availibility statement
Not applicable.
Abbreviations
- ANN:
-
Artificial neural network
- ANOVA:
-
Analysis of variance
- BFGS:
-
Broyden–Fletcher–Goldfarb–Shanno
- CRF:
-
Conditional random field
- EHR:
-
Electronic health record
- EPR:
-
Electronic patient records
- FN:
-
False negative
- FP:
-
False positive
- GRU:
-
Gated recurrent unit
- HIPAA:
-
Health Insurance Portability and Accountability Act
- HMM:
-
Hidden Markov model
- HSD:
-
Honestly significant difference
- IoT:
-
Internet of Things
- LSTM:
-
Long short-term memory
- MIRA:
-
Margin improved relaxed algorithm
- NLP:
-
Natural language processing
- NSF:
-
National Science Foundation
- PHI:
-
Protected health information
- RNN:
-
Recurrent neural network
- SVM:
-
Support vector machine
- TN:
-
True negative
- TP:
-
True positive
- UF:
-
University of Florida
References
Neamatullah I, Douglass MM, Li-wei HL, Reisner A, Villarroel M, Long WJ, Szolovits P, Moody GB, Mark RG, Clifford GD. Automated de-identification of free-text medical records. BMC Med Inf Decis Making. 2008;8(1):32.
Office for Civil Rights. : Standards for privacy of individually identifiable health information. Final rule. Federal Regis. 2002;67(157):53181.
HIPAA Journal: What is considered PHI under HIPAA. https://www.hipaajournal.com/considered-phi-hipaa/.
HIPAA Journal: De-identification of protected health information: how to anonymize PHI. https://www.hipaajournal.com/de-identification-protected-health-information/.
Portability I, Act A. Guidance regarding methods for de-identification of protected health information in accordance with the health insurance portability and accountability act (HIPAA) privacy rule 2012.
Meystre SM, Friedlin FJ, South BR, Shen S, Samore MH. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med Res Methodol. 2010;10
Kushida CA, Nichols DA, Jadrnicek R, Miller R, Walsh JK, Griffin K. Strategies for de-identification and anonymization of electronic health record data for use in multicenter research studies. Med Care. 2012;50(Suppl):S82.
Scheurwegs E, Luyckx K, Van der Schueren F, Van den Bulcke T. De-identification of clinical free text in Dutch with limited training data: a case study. Proc Workshop NLP Med Biol Assoc RANLP. 2013;2013:18–23.
Patil HK, Seshadri R. Big data security and privacy issues in healthcare. In: 2014 IEEE international congress on big data. New York: IEEE; 2014. p. 762–5.
Ferrández O, South BR, Shen S, Friedlin FJ, Samore MH, Meystre SM. Evaluating current automatic de-identification methods with veteran’s health administration clinical documents. BMC Med Res Methodol. 2012;12(1):109.
Leevy JL, Khoshgoftaar TM, Bauder RA, Seliya N. A survey on addressing high-class imbalance in big data. J Big Data. 2018;5(1):42.
Lesley WS. Risks and opportunities of data mining the electronic medical record. Phys Leadership J. 2015;2(4):40.
Yogarajan V, Pfahringer B, Mayo M. A review of automatic end-to-end de-identification: Is high accuracy the only metric? Appl Artif Intell. 2020;34(3):251–69.
Meyer H, Reudenbach C, Hengl T, Katurji M, Nauss T. How to detect and avoid overfitting in spatio-temporal machine learning applications. In: EGU general assembly conference abstracts, vol. 20, 2018. p. 8365.
Yao X. Evolving artificial neural networks. Proc IEEE. 1999;87(9):1423–47.
De Mulder W, Bethard S, Moens MF. A survey on the application of recurrent neural networks to statistical language modeling. Comput Speech Lang. 2015;30(1):61–98.
Kuan CM, Liu T. Forecasting exchange rates using feedforward and recurrent neural networks. J Appl Econom. 1995;10(4):347–64.
Sak H, Senior A, Beaufays F. Long short-term memory based recurrent neural network architectures for large vocabulary speech recognition 2014. arXiv preprint arXiv:1402.1128.
Yu Y, Si X, Hu C, Zhang J. A review of recurrent neural networks: LSTM cells and network architectures. Neural Comput. 2019;31(7):1235–70.
Graves A, Schmidhuber J. Framewise phoneme classification with bidirectional LSTM and other neural network architectures. Neural Netw. 2005;18(5–6):602–10.
Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. Neural architectures for named entity recognition 2016. arXiv preprint arXiv:1603.01360.
Li C, Bao Z, Li L, Zhao Z. Exploring temporal representations by leveraging attention-based bidirectional LSTM-RNNS for multi-modal emotion recognition. Inf Process Manage. 2020;57(3):102185.
Sutton C, McCallum A. An introduction to conditional random fields. Found Trends Mach Learn. 2012;4(4):267–373.
Kupiec J. Robust part-of-speech tagging using a hidden Markov model. Comput Speech Lang. 1992;6(3):225–42.
Wallach HM. Conditional random fields: an introduction. Technical Reports (CIS); 2004. p. 22.
Seymore K, McCallum A, Rosenfeld R. Learning hidden markov model structure for information extraction. In: AAAI-99 workshop on machine learning for information extraction; 1999. p. 37–42.
Rymarczyk T, Kozłowski E, Kłosowski G, Niderla K. Logistic regression for machine learning in process tomography. Sensors. 2019;19(15):3400.
Lafferty J, McCallum A, Pereira FC. Conditional random fields: probabilistic models for segmenting and labeling sequence data 2001.
Caruana R. Multitask learning. Mach Learn. 1997;28(1):41–75.
Vemulapalli R, Tuzel O, Liu MY. Deep gaussian conditional random field network: a model-based deep network for discriminative denoising. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2016. p. 4801–9.
Ramanathan V, Wechsler H. Phishing detection and impersonated entity discovery using conditional random field and latent Dirichlet allocation. Comput Secur. 2013;34:123–39.
Joder C, Essid S, Richard G. A conditional random field framework for robust and scalable audio-to-score matching. IEEE Trans Audio Speech Lang Process. 2011;19(8):2385–97.
Dernoncourt F, Lee JY, Uzuner O, Szolovits P. De-identification of patient notes with recurrent neural networks. J Am Med Inf Assoc. 2017;24(3):596–606.
Stubbs A, Uzuner Ö. Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/uthealth corpus. J Biomed Inf. 2015;58:S20–9.
Saeed M, Villarroel M, Reisner AT, Clifford G, Lehman LW, Moody G, Heldt T, Kyaw TH, Moody B, Mark RG. Multiparameter intelligent monitoring in intensive care ii (mimic-ii): a public-access intensive care unit database. Crit Care Med. 2011;39(5):952.
Bottou L. Large-scale machine learning with stochastic gradient descent. In: Proceedings of COMPSTAT’2010. Berlin: Springer; 2010. p. 177–86.
Liu Z, Chen Y, Tang B, Wang X, Chen Q, Li H, Wang J, Deng Q, Zhu S. Automatic de-identification of electronic medical records using token-level and character-level conditional random fields. J Biomed Inf. 2015;58:S47–52.
Jiang Z, Zhao C, He B, Guan Y, Jiang J. De-identification of medical records using conditional random fields and long short-term memory networks. J Biomed Inf. 2017;75:S43–53.
Kajiyama K, Horiguchi H, Okumura T, Morita M, Kano Y. De-identifying free text of Japanese electronic health records. EMNLP. 2018;2018:65.
Kim Y, Heider P, Meystre S. Ensemble-based methods to improve de-identification of electronic health record narratives. In: AMIA annual symposium proceedings, vol. 2018, American Medical Informatics Association; 2018. p. 663.
Lee JY, Dernoncourt F, Uzuner O, Szolovits P. Feature-augmented neural networks for patient note de-identification 2016. arXiv preprint arXiv:1610.09704.
Lee K, Filannino M, Uzuner Ö. An empirical test of GRUS and deep contextualized word representations on de-identification. Stud Health Technol Inf. 2019;264:218–22.
Liu Z, Tang B, Wang X, Chen Q. De-identification of clinical notes via recurrent neural network and conditional random field. J Biomed Inf. 2017;75:S34–42.
Madan A, George AM, Singh A, Bhatia M. Redaction of protected health information in ehrs using crfs and bi-directional lstms. In: 2018 7th international conference on reliability, Infocom Technologies and Optimization (Trends and Future Directions)(ICRITO), IEEE; 2018. p. 513–7.
Richter-Pechanski P, Amr A, Katus HA, Dieterich C. Deep learning approaches outperform conventional strategies in de-identification of German medical reports. Stud Health Technol Inf. 2019;267:101–9.
Srivastava, A., Ekbal, A., Saha, S., Bhattacharyya, P., et al.: A recurrent neural network architecture for de-identifying clinical records. In: Proceedings of the 13th international conference on natural language processing. 2016. p. 188–97.
Trienes J, Trienschnigg D, Seifert C, Hiemstra D. Comparing rule-based, feature-based and deep neural methods for de-identification of dutch medical records. In: ACM health search and data mining workshop, HSDM 2020 2020.
Yang X, Lyu T, Li Q, Lee CY, Bian J, Hogan WR, Wu Y. A study of deep learning methods for de-identification of clinical notes in cross-institute settings. BMC Med Inf Decis Making. 2019;19(5):232.
Stubbs A, Filannino M, Uzuner Ö. De-identification of psychiatric intake records: overview of 2016 CEGS n-grid shared tasks track 1. J Biomed Inf. 2017;75:S4–18.
Morita M, Kano Y, Ohkuma T, Miyabe M, Aramaki E. Overview of the ntcir-10 mednlp task. In: NTCIR. Citeseer 2013.
D’Souza J, Ng V. Ensemble-based medical relation classification. In: Proceedings of COLING 2014, the 25th international conference on computational linguistics: technical papers; 2014. p. 1682–93
Kuncheva LI, Bezdek JC, Duin RP. Decision templates for multiple classifier fusion: an experimental comparison. Pattern Recognit. 2001;34(2):299–314.
Wolpert DH. Stacked generalization. Neural Netw. 1992;5(2):241–59.
Manning CD, Surdeanu M, Bauer J, Finkel JR, Bethard S, McClosky D. The stanford corenlp natural language processing toolkit. In: Proceedings of 52nd annual meeting of the association for computational linguistics: system demonstrations, 2014. p. 55–60.
Mehne SHH, Mirjalili S. Support vector machine: Applications and improvements using evolutionary algorithms. In: Evolutionary machine learning techniques. Berlin: Springer; 2020. p. 35–50.
Crammer K, Singer Y. Ultraconservative online algorithms for multiclass problems. J Mach Learn Res. 2003;3(Jan):951–91.
Kim J, Kim H, et al. Classification performance using gated recurrent unit recurrent neural network on energy disaggregation. In: 2016 international conference on machine learning and cybernetics (ICMLC), vol. 1, New York: IEEE; 2016. p. 105–10.
Sun C, Yang Z, Luo L, Wang L, Zhang Y, Lin H, Wang J. A deep learning approach with deep contextualized word representations for chemical-protein interaction extraction from biomedical literature. IEEE Access. 2019;7:151034–46.
Chiu JP, Nichols E. Named entity recognition with bidirectional LSTM-CNNS. Trans Assoc Comput Linguist. 2016;4:357–70.
Liu DC, Nocedal J. On the limited memory BFGS method for large scale optimization. Math Programm. 1989;45(1–3):503–28.
Richter-Pechanski P, Riezler S, Dieterich C. De-identification of German medical admission notes. In: GMDS; 2018. p. 165–69.
Elman JL. Finding structure in time. Cognit Sci. 1990;14(2):179–211.
Jordan MI. Serial order: A parallel distributed processing approach. In: Advances in psychology, vol. 121, Amsterdam: Elsevier; 1997. p. 471–95.
Chang JC, Lin CC. Recurrent-neural-network for language detection on twitter code-switching corpus 2014. arXiv preprint arXiv:1412.4314.
Menger V, Scheepers F, van Wijk LM, Spruit M. Deduce: a pattern matching method for automatic de-identification of Dutch medical text. Telematics Inf. 2018;35(4):727–36.
Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. In: Advances in neural information processing systems. 2013. p. 3111–9.
Joulin A, Grave E, Bojanowski P, Douze M, Jégou H, Mikolov T. Fasttext. zip: Compressing text classification models 2016. arXiv preprint arXiv:1612.03651.
Mikolov T, Grave E, Bojanowski P, Puhrsch C, Joulin A. Advances in pre-training distributed word representations 2017. arXiv preprint arXiv:1712.09405
Johnson AE, Pollard TJ, Shen L, Li-wei HL, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG. Mimic-iii, a freely accessible critical care database. Sci Data. 2016;3:160035.
Jagannatha AN, Yu H. Structured prediction models for rnn based sequence labeling in clinical text. In: Proceedings of the conference on empirical methods in natural language processing. conference on empirical methods in natural language processing, vol. 2016, NIH Public Access; 2016. p. 856.
Berg H, Dalianis H. Augmenting a de-identification system for swedish clinical text using open resources and deep learning. In: Proceedings of the Workshop on NLP and Pseudonymisation, NoDaLiDa, Turku, Finland September, vol 30; 2019. p. 2019
Bui DDA, Wyatt M, Cimino JJ. The UAB informatics institute and 2016 CEGS n-grid de-identification shared task challenge. J Biomed Inf. 2017;75:S54–61.
Bui DDA, Redden DT, Cimino JJ. Is multiclass automatic text de-identification worth the effort? Methods Inf Med. 2018;57(04):177–84.
Du L, Xia C, Deng Z, Lu G, Xia S, Ma J. A machine learning based approach to identify protected health information in Chinese clinical text. Int J Med Inf. 2018;116:24–32.
Lee HJ, Wu Y, Zhang Y, Xu J, Xu H, Roberts K. A hybrid approach to automatic de-identification of psychiatric notes. J Biomed Inf. 2017;75:S19–27.
Lee HJ, Zhang Y, Roberts K, Xu H. Leveraging existing corpora for de-identification of psychiatric notes using domain adaptation. In: AMIA annual symposium proceedings, vol. 2017, American Medical Informatics Association; 2017. p. 1070.
Phuong ND, Chau VTN, Bao HT. A hybrid semi-supervised learning approach to identifying protected health information in electronic medical records. In: Proceedings of the 10th International Conference on Ubiquitous Information Management and Communication; 2016. p. 1–8.
Dalianis H, Velupillai S. De-identifying Swedish clinical text-refinement of a gold standard and experiments with conditional random fields. J Biomed Semant. 2010;1(1):6.
Östling R. Stagger: an open-source part of speech tagger for Swedish. North Eur J Lang Technol (NEJLT). 2013;3:1–18.
Ritter A, Clark S, Etzioni O, et al. Named entity recognition in tweets: an experimental study. In: Proceedings of the conference on empirical methods in natural language processing. Association for Computational Linguistics; 2011. p. 1524–34.
De Vries H, Elliott MN, Kanouse DE, Teleki SS. Using pooled kappa to summarize interrater agreement across many items. Field Methods. 2008;20(3):272–82.
Venkateswara H, Chakraborty S, Panchanathan S. Deep-learning systems for domain adaptation in computer vision: learning transferable feature representations. IEEE Signal Process Mag. 2017;34(6):117–29.
Jiang J, Zhai C. Instance weighting for domain adaptation in nlp. In: Proceedings of the 45th annual meeting of the association of computational linguistics; 2007. p. 264–71.
Clark JH, Lavie A, Dyer C. One system, many domains: Open-domain statistical machine translation via feature augmentation 2012.
Uzuner O, Szolovits P, Kohane I. i2b2 workshop on natural language processing challenges for clinical records. In: Proceedings of the fall symposium of the American Medical Informatics Association. Washington, DC. 2006.
Bauder RA, Herland M, Khoshgoftaar TM Evaluating model predictive performance: A medicare fraud detection case study. In: 2019 IEEE 20th international conference on information reuse and integration for data science (IRI). New York: IEEE; 2019. p. 9–14.
Iversen GR, Wildt AR, Norpoth H, Norpoth HP. Analysis of variance. Sage. 1987.
Tukey JW. Comparing individual means in the analysis of variance. Biometrics. 1949;99–114.
Katal A, Wazid M, Goudar RH. Big data: issues, challenges, tools and good practices. In: 2013 Sixth international conference on contemporary computing (IC3), IEEE; 2013. p. 404–409.
Manogaran G, Thota C, Lopez D, Vijayakumar V, Abbas KM, Sundarsekar R. Big data knowledge system in healthcare. In: Internet of things and big data technologies for next generation healthcare. Springer; 2017. pp. 133–157.
Mohammadi M, Al-Fuqaha A, Sorour S, Guizani M. Deep learning for IoT big data and streaming analytics: a survey. IEEE Commun Surv Tutor. 2018;20(4):2923–60.
Gama J, Žliobaitė I, Bifet A, Pechenizkiy M, Bouchachia A. A survey on concept drift adaptation. ACM Comput Surv (CSUR). 2014;46(4):44.
Sako Z, Adibi S, Wickramasinghe N. Addressing data accuracy and information integrity in mhealth solutions using machine learning algorithms. In: Delivering superior health and wellness management with IoT and analytics. Berlin: Springer; 2020. p. 345–59.
Moreno-Torres JG, Raeder T, Alaiz-RodríGuez R, Chawla NV, Herrera F. A unifying view on dataset shift in classification. Pattern Recognit. 2012;45(1):521–30.
Chilakapati A. Concept drift and model decay in machine learning 2019. http://xplordat.com/2019/04/25/concept-drift-and-model-decay-in-machine-learning/.
Acknowledgements
We would like to thank the reviewers in the Data Mining and Machine Learning Laboratory at Florida Atlantic University. Additionally, we acknowledge partial support by the National Science Foundation (NSF) (CNS-1427536). Opinions, findings, conclusions, or recommendations in this paper are the authors’ and do not reflect the views of the NSF.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
JLL performed the literature review and drafted the manuscript. TMK worked with JLL to develop the article’s framework and focus. TMK introduced this topic to JLL. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Leevy, J.L., Khoshgoftaar, T.M. & Villanustre, F. Survey on RNN and CRF models for de-identification of medical free text. J Big Data 7, 73 (2020). https://doi.org/10.1186/s40537-020-00351-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40537-020-00351-4