Abstract
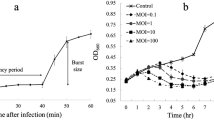
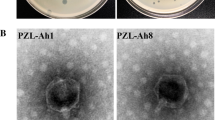
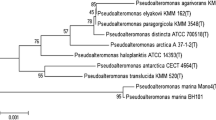
The bacteriophage vB_AhM_PVN02 (PVN02), infecting Aeromonas hydrophila, was isolated from a striped catfish pond water sample in Can Tho City, Vietnam. The phage had high lytic activity with a latent period and burst size of approximately 20 min and 105 plaque-forming units per cell, respectively. Observation of the phage by transmission electron microscopy indicated that PVN02 belongs to the family Myoviridae. The genome of PVN02 is a double-stranded linear DNA with a length in 51,668 bp and a content of 52% GC. Among the 64 genes, 16 were predicted to encode proteins with predicted functions. No virulence or antibiotic resistance genes were found in the genome, suggesting it would be a useful biocontrol agent. Classification of the phage based on sequence comparisons, phylogenetic analysis, and gene-sharing networks was carried out, and it was found to be the first representative of a new species within a previously undefined genus in the family Myoviridae. This study confirmed that PVN02 is a novel lytic phage that could potentially be used as an agent to control Aeromonas hydrophila in striped catfish in the Mekong Delta, Vietnam.


Similar content being viewed by others
References
De-Silva SS, Phuong NT (2011) Striped catfish farming in the Mekong Delta, Vietnam: a tumultuous path to a global success. Rev Aquacult 3:45–73
http://vasep.com.vn/Tin-Tuc/1200_58730/Xuat-khau-thuy-san-nam-2019-can-dich-voi-86-ty-USD.htm. Access 18 Jun 2020
Quach VCT, Tu TD, Dang PHH (2014) The current status antimicrobial resistance in Edwardsiella ictaluri and Aeromonas hydrophila cause disease on the striped catfish farmed in the Mekong Delta. J Sci Can Tho Univ Special Issue Aquac 2:7–14
Wu JL, Lin HM, Jan L, Hsu YL, Chang LH (1981) Biological control of fish bacterial pathogen, Aeromonas hydrophila, by bacteriophage AH 1. Fish Pathol 15:271–276
Richards GP (2014) Bacteriophage remediation of bacterial pathogens in aquaculture: a review of the technology. Bacteriophage. https://doi.org/10.4161/21597081.2014.975540
Doss J, Culbertson K, Hahn D, Camacho J, Barekzi N (2017) A review of phage therapy against bacterial pathogens of aquatic and terrestrial organisms. Viruses 9(3):50
Gon-Choudhury T, Tharabenahalli-Nagaraju V, Gita S, Paria A, Parhi J (2017) Advances in bacteriophage research for bacterial disease control in aquaculture. Rev Fish Sci Aquac 25(2):113–125
Hoang HA, Tran TTX, Le PN, Dang THO (2019) Selection of phages to control Aeromonas hydrophila—an infectious agent in striped catfish. Biocontrol Sci 24:23–28
Le TS, Nguyen TH, Vo HP, Doan VC, Nguyen HL, Tran MT, Tran TT, Southgate PC, Kurtböke Dİ (2018) Protective effects of bacteriophages against Aeromonas hydrophila causing motile Aeromonas Septicemia (MAS) in striped catfish. Antibiotics 7(1):16
Philipson CW, Voegtly LJ, Lueder MR, Long KA, Rice GK, Frey KG, Biswajit B, Regina ZC, Theron H, Bishop-Lilly KA (2018) Characterizing phage genomes for therapeutic applications. Viruses 10(4):188
Rai S, Tyagi A, Kalia A, Kumar BN, Garg P, Singh NK (2020) Characterization and genome sequencing of three Aeromonas hydrophila—specific phages, CF8, PS1, and PS2. Arch Virol 165:1675–1678
Panangala VS, Shoemaker CA, Van SVL, Dybvig K, Klesius PH (2007) Multiplex-PCR for simultaneous detection of 3 bacterial fish pathogens, Flavobacterium columnare, Edwardsiella ictaluri, and Aeromonas hydrophila. Dis Aquat Organ 74(3):199–208
Le HT, Truong QN, Nguyen HG, Dang THO (2010) Study on application of multiplex PCR protocol for simultaneous detection of Edwardsiella ictaluri and Aeromonas hydrophila infection in kidney of striped catfish (Pangasianodon hypophthalmus). J Sci Can Tho Univ 16a:129–135
Hoang HA, Yen MH, Ngoan VT, Nga LP, Oanh DTH (2018) Virulent bacteriophage of Edwardsiella ictaluri isolated from kidney and liver of striped catfish Pangasianodon hypophthalmus in Vietnam. Dis Aquat Organ 132(1):49–56
Verma V, Harjai K, Chhibber S (2009) Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: a potential therapeutic agent. Curr Microbiol 59:274–281
Bolger A, Giorgi F (2014) Trimmomatic: a flexible read trimming tool for illumina NGS data. Bioinformatics 30(15):2114–2120
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Christina AC, Qiandong Z, Jennifer W, Sarah KY, Earl AM (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS One 9:11
Merrill BD, Ward AT, Grose JH, Hope S (2016) Software-based analysis of bacteriophage genomes, physical ends, and packaging strategies. BMC Genomics 17(1):679
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10(1):421
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39(2):W29–W37
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780
Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32(1):268–274
Jang HB, Bolduc B, Zablocki O et al (2019) Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat Biotechnol 37:632–639
Michniewski S, Redgwell T, Grigonyte A, Rihtman B, Aguilo-Ferretjans M, Christie-Oleza J, Eleanor J, David JS, Millard AD (2019) Riding the wave of genomics to investigate aquatic coliphage diversity and activity. Environ Microbiol 21(6):2112–2128
Ackermann HW (2009) Basic phage electron microscopy. Bacteriophages. Humana press, Totowa, pp 113–126
Adriaenssens E, Brister JR (2017) How to name and classify your phage: an informal guide. Viruses 9(4):70
Alcock BP, Raphenya AR, Lau TT et al (2020) CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48(D1):D517–D525
Liu B, Zheng D, Jin Q, Chen L, Yang J (2019) VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47(D1):D687–D692
Acknowledgements
This research was funded by the Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 10/2019/TN.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: T. K. Frey.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tu, V.Q., Nguyen, TT., Tran, X.T.T. et al. Complete genome sequence of a novel lytic phage infecting Aeromonas hydrophila, an infectious agent in striped catfish (Pangasianodon hypophthalmus). Arch Virol 165, 2973–2977 (2020). https://doi.org/10.1007/s00705-020-04793-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04793-2