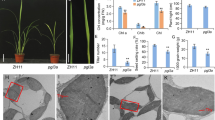
Abstract
Chloroplasts are identified as important organelles for photosynthesis, biosynthesis, and storage of metabolites. So far, the molecular regulatory mechanism of chloroplast development remains to be unclear. The chloroplast RNA splicing 2 (CRS2) protein was determined to play a key role in chloroplast development as they affect the splicing of chloroplast gene introns in maize. However, the functions of OsCRS2 in rice chloroplast development have rarely been studied. In this study, oscrs2-1 and oscrs2-2 mutants were obtained to gene function research using the CRISPR/Cas9 gene editing technology. Both of the mutants displayed an albino seedling phenotype. The OsCRS2 is located in the chloroplast, and is highly expressed in green tissues. OsCRS2 could affect group I and seven group II introns splicing and participate in chloroplast rRNA biogenesis. RNA sequencing analysis indicated that both nuclear-encoded photosynthesis and chloroplast development-related genes expression were significantly reduced in the oscrs2-1 mutant. To conclude, these results suggested that OsCRS2 is necessary for early chloroplast development in rice.








Similar content being viewed by others
References
Asakura Y, Barkan A (2006) Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol 142(4):1656–1663
Asakura Y, Barkan A (2007) A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell 19(12):3864–3875
Asakura Y, Bayraktar OA, Barkan A (2008) Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. RNA 14(11):2319–2332
Barkan A, Goldschmidt-Clermont M (2000) Participation of nuclear genes in chloroplast gene expression. Biochimie 82(6–7):559–572
Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A (2008) The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol 28(17):5337–5347
Bose J, Munns R, Shabala S, Gilliham M, Pogson B, Tyerman SD (2015) Insights into chloroplast biogenesis and development. Biochim Biophys Acta 47(9):1017–1024
Chateigner-Boutin AL, des Francs-Small CC, Delannoy E, Kahlau S, Tanz SK, de Longevialle AF, Fujii S, Small I (2011) OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. Plant J 65(4):532–542
Daras G, Rigas S, Alatzas A, Samiotaki M, Chatzopoulos D, Tsitsekian D, Papadaki V, Templalexis D, Banilas G, Athanasiadou AM, Kostourou V, Panayotou G, Hatzopoulos P (2019) LEFKOTHEA regulates nuclear and chloroplast mRNA splicing in plants. Dev Cell 50(6):767–779
de Longevialle AF, Small ID, Lurin C (2010) Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol Plant 3(4):691–705
Gong X, Jiang Q, Xu J, Zhang J, Teng S, Lin D, Dong Y (2013) Disruption of the rice plastid ribosomal protein s20 leads to chloroplast developmental defects and seedling lethality. Genes Genomes Genet 23(10):1769–1777
Gothandam KM, Kim ES, Cho H, Chung YY (2005) OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis. Plant Mol Biol 58(3):421–433
Hiei Y, Komari T (2008) Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc 3(5):824–834
Jenkins BD, Barkan A (2001) Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J 20(4):872–879
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:480–484
Kirchhoff H (2019) Chloroplast ultrastructure in plants. New Phytol 223(2):565–574
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148:350–382
Liu C, Zhu H, Xing Y, Tan J, Chen X, Zhang J, Peng H, Xie Q, Zhang Z (2016) Albino leaf 2 is involved in the splicing of chloroplast group I and II introns in rice. J Exp Bot 67(18):5339–5347
Liu X, Lan J, Huang Y, Cao P, Zhou C, Ren Y, He N, Liu S, Tian Y, Nguyen T, Jiang L, Wan J (2018) WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J Exp Bot 69(16):3949–3961
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆CT method. Methods 25:402–408
Ostersetzer O, Cooke AM, Watkins KP, Barkan A (2005) CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell 17(1):241–255
Ostheimer GJ, Williams-Carrier R, Belcher S, Osborne E, Gierke J, Barkan A (2003) Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J 22(15):3919–3929
Ostheimer GJ, Rojas M, Hadjivassiliou H, Barkan A (2006) Formation of the CRS2-CAF2 group II intron splicing complex is mediated by a 22-amino acid motif in the COOH-terminal region of CAF2. J Biol Chem 281(8):4732–4738
Robles P, Quesada V (2019) Transcriptional and post-transcriptional regulation of organellar gene expression (OGE) and its roles in plant salt tolerance. Int J Mol Sci 20(5):1056
Strand A, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421(6918):79–83
Sun Y, Tian Y, Cheng S, Wang Y, Hao Y, Zhu J, Zhu X, Zhang Y, Yu M, Lei J, Bao X, Wu H, Wang Y, Wan J (2019) WSL6 encoding an Era-type GTP-binding protein is essential for chloroplast development in rice. Plant Mol Biol 100(6):635–645
Surpin M, Larkin RM, Chory J (2002) Signal transduction between the chloroplast and the nucleus. Plant Cell 14(Suppl):S327–S338
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11(2):R14
Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, Wang J, Wang H (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7(1):30
Zhang YY, Hao YY, Wang YH, Wang CM, Wang YL, Long WH, Wang D, Liu X, Jiang L, Wan JM (2017) Lethal albinic seedling, encoding a threonyl-tRNA synthetase, is involved in development of plastid protein synthesis system in rice. Plant Cell Rep 36(7):1053–1064
Zhang Q, Shen L, Wang Z, Hu G, Ren D, Hu J, Zhu L, Gao Z, Zhang G, Guo L, Zeng D, Qian Q (2019) OsCAF1, a CRM domain containing protein, influences chloroplast development. Int J Mol Sci 20(18):4386
Zhang Q, Shen L, Ren D, Hu J, Zhu L, Gao Z, Zhang G, Guo L, Zeng D, Qian Q (2020) Characterization of the CRM gene family and elucidating the function of OsCFM2 in rice. Biomolecules 10(2):327
Zheng H, Wang Z, Tian Y, Liu L, Lv F, Kong W, Bai W, Wang P, Wang C, Yu X, Liu X, Jiang L, Zhao Z, Wan J (2019) Rice albino 1, encoding a glycyl-tRNA synthetase, is involved in chloroplast development and establishment of the plastidic ribosome system in rice. Plant Physiol Biochem 139:495–503
Acknowledgements
This research was supported by the ‘Collaborative Innovation Project’ of the Chinese Academy of Agricultural Sciences, the Central Public-interest Scientific Institution Basal Research Fund of China National Rice Research Institute (2017RG001-4), and the National Natural Science Foundation of China (31801333).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Q., Wang, Y., Shen, L. et al. OsCRS2 encoding a peptidyl-tRNA hydrolase protein is essential for chloroplast development in rice. Plant Growth Regul 92, 535–545 (2020). https://doi.org/10.1007/s10725-020-00655-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-020-00655-8