Abstract
Enhancer of zester homolog 2 (EZH2), a histone lysine methyltransferase and the catalytic component of polycomb repressive complex 2, has been extensively investigated as a chromatin regulator and a transcriptional suppressor by methylating H3 at lysine 27 (H3K27). EZH2 is upregulated or mutated in most cancers, and its expression levels are negatively associated with clinical outcomes. However, the current developed small-molecule inhibitors targeting EZH2 enzymatic activities could not inhibit the growth and progression of solid tumors. Here, we discovered an antihistamine drug, ebastine, as a novel EZH2 inhibitor by targeting EZH2 transcription and subsequently downregulating EZH2 protein level and H3K27 trimethylation in multiple cancer cell lines at concentrations below 10 μmol/L. The inhibition of EZH2 by ebastine further impaired the progression, migration, and invasiveness of these cancer cells. Overexpression of Ezh2 wild-type and its mutant, H689A (lacking methyltransferase activity), rescued the neoplastic properties of these cancer cells after ebastine treatment, suggesting that EZH2 targeted by ebastine is independent of its enzymatic function. Next-generation RNA-sequencing analysis also revealed that C4-2 cells treated with 8 μmol/L ebastine showed a gene profiling pattern similar to EZH2-knockdown C4-2 cells, which was distinctively different from cells treated with GSK126, an EZH2 enzyme inhibitor. In addition, ebastine treatment effectively reduced tumor growth and progression, and enhanced progression-free survival in triple-negative breast cancer and drug-resistant castration-resistant prostate cancer patient-derived xenograft mice. Our data demonstrated that ebastine is a novel, safe, and potent anticancer agent for patients with advanced cancer by targeting the oncoprotein EZH2.
Introduction
The polycomb group (PcG) proteins form repressive complexes (PRC) with diverse and conserved proteins that perform their functions as crucial epigenetic modifiers and transcriptional regulators in many cellular processes, including cell differentiation, cell-cycle regulation, DNA damage repair, stem cell self-renewal, and disease development and progression (1). Enhancer of zester homolog 2 (EZH2) forms the core of PRC2 along with EED and SUZ12, and methylates histone H3 at lysine 27 (H3K27) to suppress the downstream targets (1). Many studies have reported that EZH2 overexpression enhances the proliferative properties of cancer cells, while knocking down or inhibiting EZH2 could induce apoptosis and autophagy in cancers (2). Importantly, recent evidence reveals that H3K27 plays a critical role in epigenetic regulation and cancer initiation/progression. A point mutant of lysine 27 (H3K27M) could induce pediatric high-grade gliomas (3). Nevertheless, EZH2 inhibition can still effectively reduce the growth of tumors harboring the H3K27M mutant (4), suggesting that EZH2 may perform its oncogenic functions beyond its methyltransferase activities. EZH2 cross-talks with diverse epigenetic machinery in the course of chromatin compaction and gene expression regulation (5).
We and many other groups have reported that EZH2 is a biomarker of aggressive prostate and breast cancer (6, 7). The expression levels of EZH2 are profoundly involved in the progression of prostate cancer, especially in the lethal castration-resistant status (6). While EZH2 expression is very low or undetectable in benign cells, EZH2 mRNA and protein levels are elevated in advanced cancers. EZH2 overexpression in prostate and breast cancer represents a high possibility of metastasis, high Gleason score, and adverse clinical prognosis (6, 8). In addition, many studies demonstrated a strong correlation between EZH2 expression levels and the progression of many other cancer types, including, but not limited to, lung cancer (9, 10), bladder cancer (11), endometrial cancer (12), melanoma (13), as well as nonsolid neoplasms from hematopoietic and lymphoid origins (14, 15).
In search of new treatment strategies for advanced cancers, EZH2 presents to be a promising therapeutic target. Tremendous efforts have been made to develop small-molecule inhibitors against EZH2. 3-deazaneplanocin A (DZNep) is the first identified EZH2 inhibitor that targets S-adenosyl-L-homocysteine hydrolase (SAH), a cofactor known to be required for EZH2-dependent methylation (16). DZNep decreases EZH2 protein level, but it is not an ideal EZH2 inhibitor due to its nonspecific inhibition to histone methylation and excessive toxicity in animal models (17–19). Other EZH2 inhibitors, such as GSK126, EPZ5687, EPZ6438, and EI1, specifically target the lysine methyltransferase activity of EZH2, but do not alter EZH2 expression levels (15, 20–22). Disappointingly, these EZH2 enzyme inhibitors alone only suppress the growth of lymphomas harboring EZH2 gain-of-function mutations, but not that of the solid tumors, in which EZH2 is upregulated. Hereby, we report that ebastine (23), a second-generation antagonist of the histamine H1 receptor, which has been extensively evaluated for its safety and toxicity and approved for antiallergy therapy in many European countries, could be repurposed for cancer therapy by targeting EZH2 in cancer cells.
Materials and methods
Cell lines
MDA-MB-231, PC3, DU145, MCF7, H82, H146, L1236, H526, VCaP, Jeko-1, SUM159, HDLM2, and HEK293T cells were purchased from the ATCC in 2016. C4-2 was a gift from Dr. Leland W. Chung (Cedars-Sinai Medical Center, Los Angeles, CA). All cell lines were tested and authenticated at the University of Arizona Genetic Core (Tucson, AZ) in 2017 and 2019. PC3 and MDA-MB-231 were cultured in DMEM (GenDEPOT) supplemented with 10% FBS (GenDEPOT). DU145, C4-2, MCF7, H82, H146, L1236, H526, Jeko-1, and HDLM2 were cultured in RPMI1640 (GenDEPOT) supplemented with 10% FBS (GenDEPOT). SUM159 was cultured in Ham's F-12 (Gibco) supplemented with 10% FBS. All cells were frozen at passage 2–3 and used within 20 passages after each thawing. The cells were cultured in a 37°C incubator and a humidified atmosphere with 5% CO2. All cell lines were authenticated by the University of Arizona Genetics Core (Tucson, AZ) using short tandem repeat profiling. Cell lines were Mycoplasma negative as reported by routine laboratory tests.
Reagents and antibodies
GSK126 (406228, MedKoo), ebastine (ab141928, Abcam), and carebastine (23076, Cayman Chemical) were dissolved in 100% ethanol or DMSO for cell treatment. Lipofectamine 3000 (Thermo Fisher Scientific) was used to perform the transfection of EZH2 shRNA (Sigma). The following antibodies were used for Western blot analysis: EZH2 (5246, Cell Signaling Technology), GAPDH (sc-32233, Santa Cruz Biotechnology), H3K27me3 (9733, Cell Signaling Technology), H3 (9715, Cell signaling Technology), β-Actin (A2228, Sigma), and LC3-A/B (12741, Cell Signaling Technology).
Western blotting
For protein extraction, lysates were added to 1X reducing SDS sample buffer prepared by lysis buffer and 4X reducing SDS sample buffer (BP-110R, Boston BioProducts) and heated to 95°C for 10 minutes. Protein levels were assessed by standard SDS-PAGE and transferred to polyvinylidene difluoride membranes (162-0177, Bio-Rad). Images were captured using the ChemiDoc XRS+ Molecular Imager System (Bio-Rad). Primary antibodies used in Western blotting are listed above. Blots were incubated overnight with primary antibodies at 4°C, followed by detection with Clean-Blot IP Detection Reagent (HRP, 21230, Thermo Fisher Scientific), goat anti-mouse IgG (H+L)-HRP (SA001-500, GenDEPOT), or goat anti-rabbit IgG (H+L)-HRP (SA002-500, GenDEPOT) secondary antibody.
Lentiviral constructs
Lentivirus was packaged by cotransfection of constructs with third-generation packaging plasmids pMD2.G, pRRE, and pRSV/REV with Fugene HD (Roche) into HEK293T cells. The transfection mixture was replaced with growth medium 24 hours after transfection (2 μg of MDLG, 1 μg of VSVG, 1 μg of Rev, and 4 μg of target plasmid). The supernatant was collected at 72 and 96 hours after transfection and centrifuged to remove HEK293T cells. Lentiviral titers were determined by p24 assay, in addition to functional titration to determine a multiplicity of infection of 1 for each initial batch of virus.
Reporter luciferase assays
The EZH2 reporter, the void vector control, and secreted embryonic alkaline phosphatase (SEAP) plasmid construct were purchased from GeneCopoeia. Upon cultivation of C4-2 cells in 96-well plates for 24 hours, the EZH2 reporter or the void vector control was cotransfected together with the SEAP plasmid construct, which was used as an indicator of transfection internal control, into the cells. After 24 hours of incubation, the transfection system was removed and the cells were treated with 4 and 8 μmol/L of ebastine. Cells were then lysed after 24, 48, and 72 hours of incubation, and the bioluminescence test was conducted using the Secrete-Pair Dual Luminescence Assay Kit (LF061, GeneCopoeia). The bioluminescence was read on Synergy 2 Multi-Mode Reader (BioTek). EZH2 promoter luciferase activity was normalized with the void vector and SEAP luciferase activity. Each experiment was performed in quadruplicate.
RNA-sequencing analysis
The vector control, GSK126, and EZH2KD#1 and EZH2KD#2 RNA-sequencing data were obtained from a previous publication (24). Ebastine-treated samples were prepared together with control, GSK126-treated, and EZH2-knockdown (KD) samples at the same batch and sequenced at same batch. The RNA-sequencing reads were mapped to the human reference genome version hg19 using TopHat (version 2.0.12) default parameters (25). The human reference gene set (RefSeq gene) was downloaded from https://www.ncbi.nlm.nih.gov/refseq/rsg/. Cuffdiff (v2.0.12) was used to calculate gene expression levels and the significance of differential expression based on the classic FPKM using default parameters (.26). Differentially expressed gene threshold was set for P < 0.05. For clustering analysis, we used a hierarchical clustering method with Spearman correlation distance to cluster samples on the basis of the log-scaled FPKM, and used MORPHEUS (https://software.broadinstitute.org/morpheus/) to plot the heatmap. The significance of overlapping genes between the two groups was calculated using Fisher exact test. Gene set enrichment analysis (GSEA) was applied to assess the significance of associations between androgen receptor (AR) target genes and genes affected by ebastine treatment or EZH2 KD (27).
Cell growth assay
Cells were seeded in 96-well plates and treated with different concentrations of ebastine for 72 hours. Bioluminescence was measured to quantify cell viability using CellTiter-Glo Luminescent Cell Viability Assay Kit (Promega) and was read on Synergy 2 Multi-Mode Reader (BioTek). The cell proliferation curve was drawn and fit by the bioluminescence to drug concentration. IC50 was calculated with nonlinear fitting.
Wound-healing assay
Cell migrating capabilities were detected using wound-healing assay. C4-2 cells were plated with 80%–90% confluence in 6-well plates. Wounds were created across the monolayer of cell culture using a Bioclean pipette tip. The cells were incubated in serum-free medium supplemented with DMSO, 4 μmol/L, or 8 μmol/L of ebastine after rinsing with PBS. Wound closures were captured at 0, 24, and 72 hours.
Boyden chamber invasion assay
Polycarbonate Membrane Cell Culture Inserts (CLS3422, Corning) were applied with Basement Membrane Matrix (Trevigen). After the matrix condensed at 37°C in cell incubator, each 24-well insert was seeded with 1 × 105 of C4-2 cells in RPMI1640 without FBS. RPMI1640 with FBS was added to each well containing the insert. Ebastine or vehicle (ethanol or DMSO) was added to keep the same concentration inside and outside of the inserts. Each insert was fixed with methanol, and cells that permeated through the membrane were stained with 0.5% crystal violet. Images were captured, and the cell count was calculated using ImageJ (1.8.0).
EZH2 rescue assay
Ezh2 wild-type and Ezh2-mutant H689A cloned into MigR1 (GFP) retroviral vector were provided by Dr. Yi Zhang (Temple University, Philadelphia, PA). Retrovirus was packaged by cotransfection of constructs with second-generation packaging plasmids pUMVC and pCMV-VSVG with Lipofectamine (Thermo Fisher Scientific) into HEK293T cells. The transfection mixture was replaced with growth medium 24 hours after transfection (1 μg of pCMV-VSVG, 3 μg of pUMVC, and 4 μg of target plasmid). The supernatant was collected at 72 and 96 hours after transfection and centrifuged to remove the cells. For retroviral infection, retrovirus supernatant was added to C4-2 cells in the presence of 10 μg/mL Polybrene (Sigma). Cells were incubated at 37°C for 6–7 hours, and then transferred to fresh growth medium. The same retroviral infection procedure was repeated the next day. The proliferative, invasive, and migrating properties of these cells in response to ebastine treatment were evaluated as described in previous sections.
Autophagy and apoptosis assays
C4-2 cells were treated with ebastine at dose gradients for 72 hours and lysed for Western blotting to detect LC3-A/B. Densitometry measurements of bands were quantitated and calculated in ImageJ (1.8.0). Autophagosome activity was detected with specific dye using an Autophagy Assay Kit (MAK138, Sigma). The pictures were captured under fluorescence microscopy, and bioluminescence was read on Synergy 2 Multi-Mode Reader (BioTek).
Apoptosis was detected using FITC Annexin V Apoptosis Detection Kit (556547, BD Biosciences) in C4-2 cells treated with ebastine for 72 hours. The staining was analyzed by Flow Cytometry (LX200 Luminex Multiplexing Assay System).
Murine prostate and breast tumor xenograft models
CB17SCID mice were purchased from Charles River Laboratories. Animal care and conditions were based on the institutional and NIH protocols and guidelines, and all studies were approved by Houston Methodist Institution Animal Care and Use Committee. Tumor xenograft model was induced as described previously (28).
For VCaP castration-resistant prostate tumor model, male mice were anesthetized using 2% isoflurane (inhalation), and 2 × 106 of VCaP prostate cancer cells suspended in 100 μL of PBS with 50% Basement Membrane Matrix (Trevigen) were implanted subcutaneously into the dorsal flank on the right side of each mouse. Tumor volumes were measured by length (a) and width (b), and calculated as tumor volume = MIN (a)2 × MAX (b)/2. For VCaP castration-resistant prostate tumor model, VCaP tumor-bearing mice were castrated when tumors were approximately 200–300 mm3 in size (∼5 weeks after implantation of tumor cells), and once tumors started to relapse, mice were randomized and treated with vehicle (n = 13) or ebastine (10 mg/kg/day, n = 10, 5 days/week) orally. All animals were euthanized after 28 days of treatment. The body weight of the mice was monitored during the study.
For SUM159 triple-negative breast tumor model, female mice were anesthetized using 2% isoflurane (inhalation), and 2 × 106 of SUM159 prostate cancer cells suspended in 100 μL of PBS with 50% Basement Membrane Matrix (Trevigen) were implanted subcutaneously into the mammary fat pad. Tumor volumes were measured by length (a) and width (b), and calculated as tumor volume = MIN (a)2 × MAX (b)/2. Mice were randomized and treated with vehicle (n = 11) or ebastine (30 mg/kg/day, n = 8, 5 days/week) via oral gavage, and terminated about 28 days later. Body weight of mice was monitored throughout the study.
Murine prostate and breast patient-derived xenograft models
Mice with LuCaP 35CR castration-resistant prostate cancer (CRPC) patient-derived xenograft [PDX; gift from Dr. Eva Corey (University of Washington, Seattle, WA)] or BCM3887 triple-negative breast cancer (TNBC) PDX (gift from Dr. Jenny C. Chang) were euthanized when the tumor size more than 1,000 mm3. All cells used in this model were tested for murine-associated pathogens at IDEXX BioResearch. The tumors were excised and minced in wash medium (DMEM with 5% FBS and 50 μg/mL gentamicin). One piece of minced PDX tumor (about 8 mm3) was implanted subcutaneously into the dorsal flank on the right side of the 5-week-old precastrated male mice (LuCaP 35CR model) or 5-week-old female mice (BCM3887). For the BCM3887 group, when the tumors were approximately 100 mm3 in size, mice were randomized and treated with vehicle (n = 10) or ebastine (10 mg/kg/day, n = 5 or 30 mg/kg/day, n = 5, 5 days/week). For the LuCaP 35CR group, when the tumors were approximately 100 mm3 in size, mice were randomized and treated with enzalutamide only (10 mg/kg/day, n = 12, 5 days/week) or ebastine + enzalutamide (30 and 10 mg/kg/day, respectively, n = 13, 5 days/week). Tumor diameters and body weights were recorded twice per week. Mice were euthanized about 28 days later.
Quantification of ebastine and its metabolite, carebastine, in mouse tumor
Ebastine and carebastine in mouse tumors were extracted and measured on the basis of a previously established method with slight modifications (23). An internal standard, methaqualone (10 μmol/L in acetonitrile), and external standard, terfenadine (2.5 μg/mL in acetonitrile), were obtained from Sigma (M-015) and Tocris Bioscience (3948/50), respectively. Briefly, each tumor sample was weighed and homogenized in five times its volume of phosphate buffer solution (0.5 mol/L, pH 7.4) on ice using a Sonicator Ultrasonic Homogenizer (EpiShear Probe Sonicator, Active Motif). Acetonitrile with methaqualone was added to the homogenate, vortexed vigorously for 1 minute, and centrifuged at 15,700 × g for 10 minutes to precipitate proteins from the homogenate. The supernatant was collected for quantification.
Standards and samples were both spiked with an equal amount of internal standards and analyzed by high-performance liquid chromatography and triple-quadruple mass spectrometry and tandem mass spectrometry (HPLC/MS-MS). Specifically, the system consisted of a Thermo TSQ in-line with an electrospray source and a Vanquish (Thermo Fisher Scientific) UHPLC consisting of a binary pump, degasser, and autosampler outfitted with a Kinetex EVO C18 Column (Phenomenex, dimensions of 2.1 mm × 50 mm and a 5 μm particle size). Elution was at isocratic mode with a mobile phase containing 5 mmol/L ammonium acetate in water and acetonitrile (50%:50%, v/v) at 0.4 mL/minute. In positive mode, the capillary of electrospray source was set to 350°C, with sheath gas at 50 arbitrary units, auxiliary gas at 10 arbitrary units, and the spray voltage at 3.5 kV. A selective reaction monitoring of the protonated precursor ion and the related product ions for ebastine, carebastine, terfenadine, and methaqualone (m/z 470.2 → 167.1, 500.2 → 191.1, 472.3 → 436.1, and 251.4 → 132, respectively) was monitored. Calibration graphs were derived from the peak area ratio of targets to internal standard with linear regression. Data acquisition and analysis were carried out by Xcalibur 4.1 Software and Tracefinder 4.1 Software, respectively (both from Thermo Fisher Scientific).
Results
Ebastine decreases EZH2 and H3K27 methylation levels in cancer
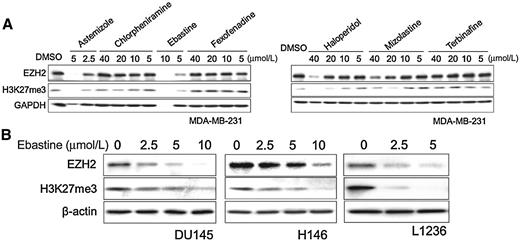
Recently, several antihistamine drugs have been reported to effectively inhibit tumor growth (29). Among these drugs, astemizole has been shown to disrupt EZH2–EED interaction and induce EZH2 degradation (30). Unfortunately, the use of astemizole, particularly overdose, may cause ventricular arrhythmias (31). Other antihistamine drugs were screened to determine the potential to decrease EZH2 protein levels in cancer cells. Besides astemizole, ebastine markedly reduced EZH2 and H3K27 trimethylation levels at concentrations below 20 μmol/L as compared with other antihistamine drugs, including chlorpheniramine, fexofenadine, haloperidol, mizolastine, or terbinafine, in MDA-MB-231 (Fig. 1A) and MCF7 (Supplementary Fig. S1A) breast cancer cells. To further investigate whether ebastine can reduce EZH2 in other cancer cells, we treated prostate cancer cells (DU145 and VCaP), small-cell lung cancer (H146, H82, and H526), and lymphoma (L1236, Jeko-1, and HDLM2) with ebastine at various concentrations. As shown in Fig. 1B; Supplementary Fig. S1B, ebastine dose dependently reduced the protein level of EZH2 and H3K27me3 in these cancer cell lines.
Ebastine decreases EZH2 protein and H3K27 methylation in cancer. A, MDA-MB-231 cells were treated with astemizole, chlorpheniramine, ebastine, fexofenadine, and haloperidol at different doses. After 72 hours, total cell lysates were blotted for EZH2 and H3K27me3, with GAPDH as a loading control. B, DU145, H146, and L126 cells were treated with ebastine at different doses for 72 hours and lysed for immunoblot analysis. β-actin served as a loading control.
Ebastine decreases EZH2 protein and H3K27 methylation in cancer. A, MDA-MB-231 cells were treated with astemizole, chlorpheniramine, ebastine, fexofenadine, and haloperidol at different doses. After 72 hours, total cell lysates were blotted for EZH2 and H3K27me3, with GAPDH as a loading control. B, DU145, H146, and L126 cells were treated with ebastine at different doses for 72 hours and lysed for immunoblot analysis. β-actin served as a loading control.
Ebastine decreased EZH2 transcript levels by regulating its promoter activities
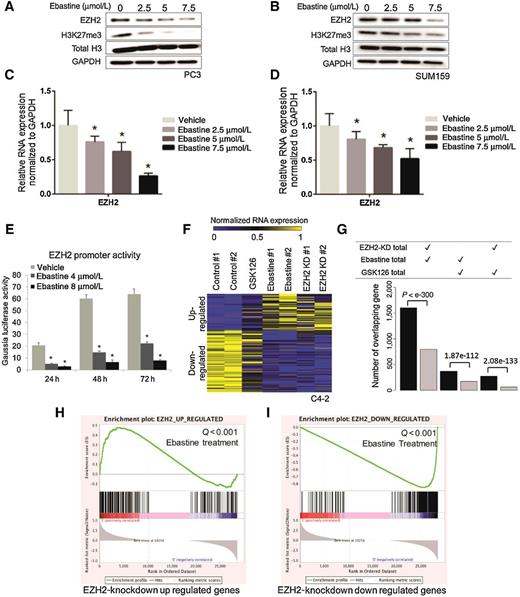
The decrease of EZH2 protein may be a result of either increased EZH2 degradation or inhibited transcription or translation. Real-time quantitative PCR (RT-qPCR) analysis demonstrated that ebastine markedly reduced the mRNA level of EZH2 dose dependently along with the reduction of EZH2 protein level in PC3 (Fig. 2A and C), C4-2 (Supplementary Fig. S2A and S2B), and SUM159 (Fig. 2B and D). Interestingly, the protein level of EED, but not SUZ12, decreased along with EZH2 after treatment with ebastine (Supplementary Fig. S2C), which is consistent with our previous finding (24). We then hypothesized that ebastine may repress EZH2 transcription by inhibiting its promoter activities. To test this hypothesis, we cotransfected a secreted Gaussia luciferase reporter containing EZH2 upstream promoter, along with a SEAP reporter as an internal control for transfection efficiency into C4-2 cells. We observed that the promoter activities of EZH2 significantly declined with ebastine treatment in a dose-dependent manner (Fig. 2E). Moreover, we conducted a next-generation RNA-sequencing in C4-2 cells treated with ebastine (8 μmol/L), GSK126 (8 μmol/L, EZH2 enzymatic inhibitor), and EZH2 shRNA (EZH2 KD). Intriguingly, we observed a similar gene expression pattern (Supplementary Fig. S3A) and a considerable overlapping of upregulated or downregulated genes in ebastine-treated cells and EZH2-KD cells, whereas the GSK126-treated group showed a distinct gene profiling pattern as compared with EZH2 KD (Fig. 2F and G). GSEA further confirmed that ebastine treatment–increased genes were significantly enriched (Q value < 0.001) in the signature of EZH2-KD upregulated genes (Fig. 2G), and ebastine treatment–decreased genes were significantly enriched (Q value < 0.001) in the signature of EZH2-KD downregulated genes (Fig. 2H). Among all of the cancer-related pathways, cell cycle was significantly downregulated after ebastine treatment (Supplementary Fig. S3B and S3C). Our results demonstrate that ebastine treatment decreased EZH2 transcript in cancer cells, and exhibited similar effect as EZH2 KD in C4-2 cells, suggesting that ebastine may be a better inhibitor for EZH2 as compared with GSK126.
Ebastine decreases EZH2 transcript levels by regulating EZH2 promoter activities. PC3 cells (A) and SUM159 (B) treated with ebastine for 72 hours were lysed and blotted for EZH2 and H3K27me3, with H3 and GAPDH as a loading control. RNA was extracted from PC3 cells (C) and SUM159 (D) treated with ebastine for 72 hours and quantified using RT-qPCR. The relative expression of EZH2 was normalized by GAPDH. *, P < 0.05 versus vehicle, mean ± SEM. E, C4-2 cells were cotransfected with a secreted Gaussia luciferase reporter containing EZH2 upstream promoter and a SEAP reporter. The SEAP group acted as an internal control. Ebastine treatment was performed for 24, 48, or 72 hours. F, Heatmap for the expression level of genes down- or upregulated by EZH2 KD, GSK126 (8 μmol/L), or ebastine (8 μmol/L) treatment. G, The number of overlapped differential genes between EZH2-KD and ebastine-treated group was significantly larger than the number of genes overlapped by GSK126-treated versus EZH2-KD or ebastine-treated group. GSEA of ebastine-targeted genes that were significantly enriched by genes upregulated after EZH2 KD (Q value < 0.001; H) or downregulated after EZH2 KD (Q value < 0.001; I).
Ebastine decreases EZH2 transcript levels by regulating EZH2 promoter activities. PC3 cells (A) and SUM159 (B) treated with ebastine for 72 hours were lysed and blotted for EZH2 and H3K27me3, with H3 and GAPDH as a loading control. RNA was extracted from PC3 cells (C) and SUM159 (D) treated with ebastine for 72 hours and quantified using RT-qPCR. The relative expression of EZH2 was normalized by GAPDH. *, P < 0.05 versus vehicle, mean ± SEM. E, C4-2 cells were cotransfected with a secreted Gaussia luciferase reporter containing EZH2 upstream promoter and a SEAP reporter. The SEAP group acted as an internal control. Ebastine treatment was performed for 24, 48, or 72 hours. F, Heatmap for the expression level of genes down- or upregulated by EZH2 KD, GSK126 (8 μmol/L), or ebastine (8 μmol/L) treatment. G, The number of overlapped differential genes between EZH2-KD and ebastine-treated group was significantly larger than the number of genes overlapped by GSK126-treated versus EZH2-KD or ebastine-treated group. GSEA of ebastine-targeted genes that were significantly enriched by genes upregulated after EZH2 KD (Q value < 0.001; H) or downregulated after EZH2 KD (Q value < 0.001; I).
Ebastine impaired malignant neoplasm and induced cell-cycle arrest and autophagy
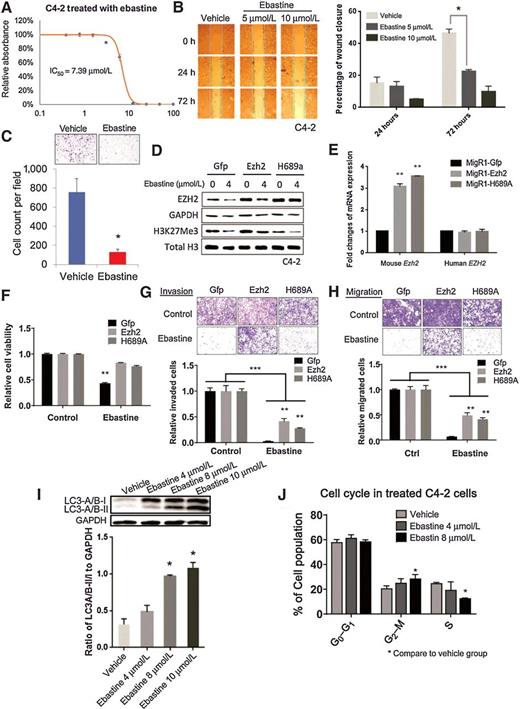
The mRNA and protein levels of EZH2 are associated with the neoplastic properties in many cancers (6, 8–14). Consistently, we found that ebastine treatment significantly inhibited the growth of various cancer cell lines with IC50 below 12 μmol/L (Fig. 3A; Supplementary Fig. S4A–S4G). In addition, ebastine treatment dose dependently abolished the ability to migrate and invade in C4-2 cells, as demonstrated by wound-healing assay (Fig. 3B) and Boyden chamber invasion assay (Fig. 3C), respectively. Similar results were observed in MDA-MB-231 breast cancer cells (Supplementary Fig. S4H). To confirm the antineoplastic activity of ebastine is specifically targeting EZH2, we overexpressed Ezh2 wild-type and its H689A mutant (lacking methyltransferase activity) driven by a MSCV promoter in C4-2 cells and NIH3T3 cells. We found that wild-type Ezh2 restored EZH2 and H3K27 protein levels, while H689A mutant only restored EZH2 protein levels, but not H3K27me levels in C4-2 cells (Fig 3D and E) and NIH3T3 cells (Supplementary Fig. S5A). Interestingly, both Ezh2 wild-type and H689A mutant could significantly rescue ebastine treatment–reduced neoplastic properties of C4-2 prostate cancer cells (Fig. 3F–H) as well as NIH3T3 cells (Supplementary Fig. S5B), suggesting that the inhibition of cancerous properties by ebastine is through targeting EZH2 regardless of its enzymatic function.
Ebastine has potent therapeutic effects on prostate cancer. A, Growth curve of C4-2 cells treated with ebastine at different concentrations to determine the IC50. B, Migration images of C4-2 cells treated with ebastine and vehicle at 0, 24, and 72 hours. The percentage of closure was calculated by dividing the width of 0 hour. *, P < 0.05 versus vehicle, mean ± SEM. C, Invasion assay using transwell insert and Matrigel to assess the invasiveness of C4-2 cells after ebastine treatment. Cell count was analyzed in the bottom panel. **, P < 0.01 versus vehicle, mean ± SEM. EZH2 rescue assay in C4-2 cells for protein level (D), mRNA level (E), growth (F), invasion (G), and migration (H). I, C4-2 cells treated with ebastine for 48 hours were lysed and blotted with anti-LC3-A/B antibody, and presented as the ratio of LC3-A/B-II/I to GAPDH. J, Cell-cycle distribution of C4-2 cells treated with ebastine and vehicle.
Ebastine has potent therapeutic effects on prostate cancer. A, Growth curve of C4-2 cells treated with ebastine at different concentrations to determine the IC50. B, Migration images of C4-2 cells treated with ebastine and vehicle at 0, 24, and 72 hours. The percentage of closure was calculated by dividing the width of 0 hour. *, P < 0.05 versus vehicle, mean ± SEM. C, Invasion assay using transwell insert and Matrigel to assess the invasiveness of C4-2 cells after ebastine treatment. Cell count was analyzed in the bottom panel. **, P < 0.01 versus vehicle, mean ± SEM. EZH2 rescue assay in C4-2 cells for protein level (D), mRNA level (E), growth (F), invasion (G), and migration (H). I, C4-2 cells treated with ebastine for 48 hours were lysed and blotted with anti-LC3-A/B antibody, and presented as the ratio of LC3-A/B-II/I to GAPDH. J, Cell-cycle distribution of C4-2 cells treated with ebastine and vehicle.
EZH2 is involved in many physiologic and pathologic cellular processes as an epigenetic regulator (32). A previous study reported that knocking down EZH2 may induce cell-cycle arrest and autophagy (32, 33). Consistently, we found that ebastine treatment induced autophagy dose dependently as indicated by the elevated ratio of LC3-A/B-II to LC3-A/B-I using Western blotting (Fig. 3I) and increased staining of proprietary fluorescence autophagosome (Supplementary Fig. S4I) in C4-2 cells. Furthermore, ebastine induced G2–M-phase cell-cycle arrest in a dose-dependent manner (Fig. 3J). Our data clearly demonstrate that ebastine inhibits the oncogenic phenotypes of cancer cells in vitro by targeting EZH2 and induces cell-cycle arrest and autophagy.
To compare the effects of ebastine and EZH2/PRC2 enzymatic inhibitors in inhibiting cancer cell growth, migration, and invasion, we treated Du145 and C4-2 cells with 4 μmol/L of ebastine, GSK126, EPZ6438, and EED226 in parallel for 72 hours. Immunoblot analysis revealed that all drugs decreased H3K27me3 levels, but only ebastine decreased EZH2 protein levels (Supplementary Fig. S6A and S6B). While the GSK126, EPZ6438, and EED226 inhibited Du145 cell growth with IC50 of 20.18 μmol/L, 77.58 μmol/L, and 24.62 μmol/L, respectively, ebastine showed a better inhibition in cell growth with IC50 of 11.15 μmol/L (Supplementary Fig. S6C). A similar effect was observed in C4-2 cells (Supplementary Fig. S6D). In addition, the Boyden chamber invasion assay and transwell migration assays revealed that ebastine significantly and markedly reduced the migration and invasive properties of Du145 and C4-2 (Supplementary Fig. S6E and S6F), while GSK126, EPZ6438, and EED226 only marginally reduced these properties of cancer cells.
Ebastine inhibits tumor progression in vivo
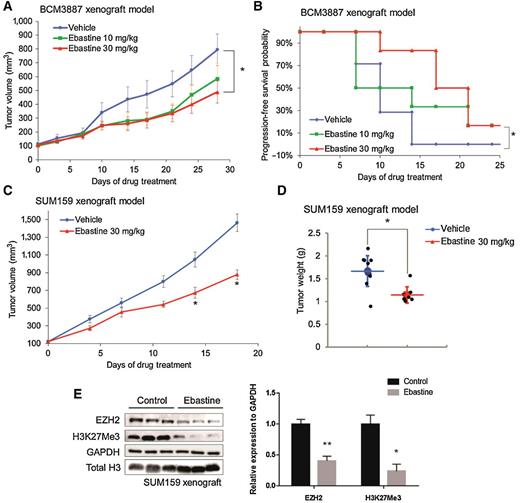
We then evaluated the therapeutic potential of ebastine against TNBC in vivo using murine xenograft models with TNBC PDX, BCM3887, and TNBC cell line, SUM159 xenograft. Once tumor size reached to 100 mm3, mice were randomized into three groups (vehicle, ebastine 10 mg/kg/day, and ebastine 30 mg/kg/day). As shown in Fig. 4A, ebastine treatment significantly and dose dependently reduced tumor growth and progression as indicated by tumor weight. Kaplan–Meier survival plot indicated that a higher dose of ebastine enhanced the survival of TNBC PDX mice (Fig. 4B). Similarly, ebastine treatment reduced tumor growth and progression in SUM159 xenograft mice, as indicated by tumor volume (Fig. 4C) and tumor weight (Fig. 4D). More importantly, immunoblot analysis using tumor lysate from the ebastine-treated and control mice showed that EZH2 protein levels were decreased in vivo upon ebastine treatment (Fig. 4E). No significant change in body weight of these mice was observed among different treatments (Supplementary Fig. S7A and S7B).
Therapeutic efficacy of ebastine in TNBC murine models. BCM3887 PDX mouse model was generated as described in the “Materials and Methods” section. Mice carrying BCM3887 PDX received vehicle or ebastine (10 and 30mg/kg/day) 5 days per week, orally. A, Caliper measurements were taken every 4 days to determine tumor volume. *, P < 0.05 versus vehicle, mean tumor volume ± SEM. B, Kaplan–Meier survival plot comparing progression-free survival of BCM3887 PDX model. Significance was tested using unpaired t test and presented as *, P < 0.05. C, Caliper measurements were taken every 4 days to determine the tumor volume of SUM159 xenograft mice. D, Tumor weight was measured after sacrifice. E, Tumor samples (n = 7) were ground and lysed for Western blot analysis. Proteins were blotted and quantitated to compare the level of EZH2 between the ebastine-treated group and the vehicle-treated group. *, P < 0.05 versus vehicle; mean ± SEM.
Therapeutic efficacy of ebastine in TNBC murine models. BCM3887 PDX mouse model was generated as described in the “Materials and Methods” section. Mice carrying BCM3887 PDX received vehicle or ebastine (10 and 30mg/kg/day) 5 days per week, orally. A, Caliper measurements were taken every 4 days to determine tumor volume. *, P < 0.05 versus vehicle, mean tumor volume ± SEM. B, Kaplan–Meier survival plot comparing progression-free survival of BCM3887 PDX model. Significance was tested using unpaired t test and presented as *, P < 0.05. C, Caliper measurements were taken every 4 days to determine the tumor volume of SUM159 xenograft mice. D, Tumor weight was measured after sacrifice. E, Tumor samples (n = 7) were ground and lysed for Western blot analysis. Proteins were blotted and quantitated to compare the level of EZH2 between the ebastine-treated group and the vehicle-treated group. *, P < 0.05 versus vehicle; mean ± SEM.
Androgen deprivation therapy (ADT) is the first-line of treatment against early-stage prostate cancer. However, most patients who receive ADT will inevitably develop castration resistance, which poses a huge challenge to clinical treatment. In the context of prostate cancer, EZH2 oncogenic activity is correlated with tumor progression and castration resistance. To evaluate the therapeutic effects of ebastine in prostate cancer, we first utilized the CRPC VCaP xenograft mouse model. As expected, ebastine treatment significantly and markedly reduced the VCaP CRPC xenograft tumor growth (Fig. 5A). We also observed better survival of mice with ebastine treatment as compared with those with vehicle control, according to Kaplan–Meier analysis (Fig. 5B). We then evaluated the therapeutic efficacy of ebastine in LuCaP 35CR, an aggressive CRPC, enzalutamide-resistant, and abiraterone-resistant PDX model (34). The combination of ebastine with enzalutamide significantly enhanced the anticancer activity of enzalutamide in the LuCaP 35CR PDX model as indicated by the inhibition of tumor growth (Fig. 5C), the reduction of overall tumor weight (Fig. 5D), and enhanced progression-free survival (Fig. 5E). Noteworthy, the protein and mRNA levels of EZH2 and its downstream targets were significantly reduced in the tumor lysate of LuCaP 35CR xenograft mice after ebastine treatment as compared with those of control (Fig. 5F and G). The levels of ebastine and its metabolite, carebastine, were measured using HPLC/MS-MS, and it was found that ebastine and carebastine levels significantly elevated 20-fold after ebastine treatment in the tumor lysate of mice (Fig. 5H and I). Again, no significant changes in body weight were observed in mice among different treatment groups (Supplementary Fig. S7C and S7D).
Therapeutic efficacy of ebastine in CRPC murine models. Castration-resistant VCaP xenograft mouse models were generated as described in the “Materials and Methods” section. Castrated mice carrying CRPC xenograft received vehicle or ebastine treatment (10 and 30 mg/kg) 5 days per week. A, Caliper measurements were taken every 4 days to obtain tumor volume. Mean tumor volume ± SEM; *, P < 0.05 versus vehicle. B, Kaplan–Meier survival plot comparing progression-free survival and significance was tested using unpaired t test. LuCap 35CR PDX mouse models were established. Mice received enzalutamide (10 mg/kg/day) or enzalutamide (10 mg/kg/day) + ebastine (30 mg/kg/day) orally. C, Tumor volume was obtained using caliper every 4 days. Mean tumor volume ± SEM. D, Tumors were dissected and weighted after 35 days of treatment. Significance was tested using unpaired t test. *, P < 0.05 versus vehicle; mean ± SEM. E, Kaplan–Meier survival plot comparing progression-free survival. Significance was tested using unpaired t test. F, Tumor samples (control, n = 6 and ebastine, n = 7) were ground and lysed for Western blot analysis. Proteins were blotted and quantitated to compare the level of EZH2, AR, and PSA between the ebastine-treated group and the vehicle-treated group. G, RNA was extracted to compare the level of EZH2, AR, and PSA. The relative expression was standardized with GAPDH. H and I, The concentration of carebastine, metabolites of ebastine, and ebastine using HPLC/MS-MS from tumor lysates. *, P < 0.05; ***, P < 0.0001 versus vehicle; mean ± SEM.
Therapeutic efficacy of ebastine in CRPC murine models. Castration-resistant VCaP xenograft mouse models were generated as described in the “Materials and Methods” section. Castrated mice carrying CRPC xenograft received vehicle or ebastine treatment (10 and 30 mg/kg) 5 days per week. A, Caliper measurements were taken every 4 days to obtain tumor volume. Mean tumor volume ± SEM; *, P < 0.05 versus vehicle. B, Kaplan–Meier survival plot comparing progression-free survival and significance was tested using unpaired t test. LuCap 35CR PDX mouse models were established. Mice received enzalutamide (10 mg/kg/day) or enzalutamide (10 mg/kg/day) + ebastine (30 mg/kg/day) orally. C, Tumor volume was obtained using caliper every 4 days. Mean tumor volume ± SEM. D, Tumors were dissected and weighted after 35 days of treatment. Significance was tested using unpaired t test. *, P < 0.05 versus vehicle; mean ± SEM. E, Kaplan–Meier survival plot comparing progression-free survival. Significance was tested using unpaired t test. F, Tumor samples (control, n = 6 and ebastine, n = 7) were ground and lysed for Western blot analysis. Proteins were blotted and quantitated to compare the level of EZH2, AR, and PSA between the ebastine-treated group and the vehicle-treated group. G, RNA was extracted to compare the level of EZH2, AR, and PSA. The relative expression was standardized with GAPDH. H and I, The concentration of carebastine, metabolites of ebastine, and ebastine using HPLC/MS-MS from tumor lysates. *, P < 0.05; ***, P < 0.0001 versus vehicle; mean ± SEM.
Discussion
EZH2 has been reported as an oncogene for more than 18 years (6, 7). The emerging role of EZH2 in cancer progression and metastasis prompts researchers and pharmaceutical companies to develop EZH2 inhibitors for cancer therapy. A number of small-molecular EZH2 inhibitors have recently been identified. Among these inhibitors, GSK126, EPZ005687, EPZ006438, and EI1 are remarkable for their high selectivity to EZH2, improved pharmacokinetic properties, and fewer side-effects (15, 20, 35). Despite their high therapeutic and clinical translation potential, the use of these EZH2 inhibitors is limited to sarcomas or lymphomas harboring EZH2 gain-of-function mutants (15, 35–38). Several studies reported that these EZH2 enzymatic inhibitors alone are not sufficient to suppress the growth and progression of solid tumors with high wild-type EZH2 expression levels unless these EZH2 inhibitors are combined with other inhibitors (39–41), or in tumors harboring other mutations such as ARID1A, EGFR, or BRG1 mutants (42, 43).
Mounting evidence suggests that EZH2 can perform its oncogenic functions independently of its methyltransferase activity (44, 45). EZH2 could act as a transcriptional activator to activate AR expression by binding to enhancer regions of AR to promote prostate cancer progression (40). EZH2 can also regulate NF-κB signaling independently of its C-terminal enzymatic SET domain by interacting with REL-A and REL-B or activating REL-B in breast cancer (46, 47). Therefore, targeting the transcript or protein levels of EZH2 could be a better therapeutic option by inhibiting both methyltransferase-dependent and -independent oncogenic functions of EZH2.
In this study, we first discovered ebastine, an antihistamine drug for allergy treatment, as a potent EZH2 inhibitor by targeting the promoter of EZH2, which downregulates both mRNA and protein levels of EZH2. It has been previously reported that the mRNA and protein levels of EZH2 are associated with the neoplastic properties in many cancers (6, 8–14). We consistently demonstrated that ebastine effectively inhibits the proliferation, migration, and invasion of various breast and prostate cells in vitro. When compared with GSK126 and other EZH2 enzyme inhibitors, EPZ6438 and EED226, ebastine showed better inhibition against EZH2 and demonstrated more robust anticancer activities. The RNA-sequencing analysis also demonstrated that cells treated with ebastine exhibited gene regulation signature as similar to EZH2 KD, which was distinct from those treated with GSK126. Most importantly, this inhibition of EZH2 by ebastine is independent of EZH2 enzymatic activity, as demonstrated by the rescue using the H689A mutant. In addition, the in vivo studies in murine xenograft models and PDX models further supported that ebastine could effectively inhibit tumor growth and progression of advanced cancers and drug-resistant cancers. In addition to ebastine, its metabolite, carebastine, was also found to be elevated in the tumor of mice treated with ebastine, and demonstrated to reduce EZH2 protein level and exhibit anticancer activities in C4-2 and DU145 prostate cancer cells (Supplementary Fig. S8).
Altogether, our results revealed that ebastine could be a promising therapeutic intervention for cancers bearing EZH2 overexpression or mutations. A retrospective study reported that patients with breast cancer taking ebastine for allergy treatment had a better overall and breast cancer–specific survival rate compared with those not taking antihistamine drugs (48–50), further suggesting the safety and the potential of using ebastine in clinical settings. With the current data, ebastine is a safe drug for daily use and should be implemented in cancer treatment intervention either as a preventive agent for people with allergy syndrome or therapeutic options for patients with advanced cancers after careful and systematic evaluation.
Disclosure of Potential Conflicts of Interest
A. Patnaik reports grants from Bristol Myers Squibb, other from Prime Inc (CME lectures), Exelixis (advisory board), Janssen (advisory board), Jounce Therapeutics (advisory board), Guidepoint (consulting), Merck (invited lecture), and Roche (invited lecture) outside the submitted work. Q. Cao reports grants from NIH, American Cancer Society, U.S. Department of Defense, and Prostate Cancer Foundation during the conduct of the study, and other from NIH (scientific reviewer compensation), U.S. Department of Defense (Scientific reviewer compensation), and American Cancer Society (scientific reviewer compensation, donated back) outside the submitted work. No potential conflicts of interest were disclosed by the other authors.
Authors' Contributions
Q. Li: Conceptualization, data curation, formal analysis, investigation, writing-original draft. K.Y. Liu: Data curation, formal analysis, investigation, methodology, writing-original draft, project administration, writing-review and editing. Q. Liu: Conceptualization, formal analysis, investigation, writing-original draft, project administration. G. Wang: Data curation, software, formal analysis, validation, investigation, methodology. W. Jiang: Resources. Q. Meng: Conceptualization, resources. Y. Yi: Conceptualization, resources, supervision. Y. Yang: Conceptualization, resources. R. Wang: Conceptualization, resources. S. Zhu: Conceptualization, resources, methodology. C. Li: Resources. L. Wu: Resources, data curation. D. Zhao: Resources. L. Yan: Resources. L. Zhang: Resources. J.-S. Kim: Conceptualization, resources. X. Zu: Resources, supervision, funding acquisition. A.J. Kozielski: Resources. W. Qian: Resources, data curation. J.C. Chang: Resources. A. Patnaik: Resources. K. Chen: Conceptualization, resources, supervision, funding acquisition, validation, investigation, project administration. Q. Cao: Conceptualization, resources, supervision, funding acquisition, validation, investigation, visualization, writing-original draft, project administration, writing-review and editing.
Acknowledgments
We would like to acknowledge Dr. Peng Gao at the Metabolomics Developing Core Facility of Northwestern University for the HPLC/MS-MS support. We also wish to acknowledge the following individuals for their kind gifts: Dr. Leland W. Chang for the C4-2 cells, Dr. Yi Zhang for the MigR1 (GFP) retroviral vectors of Ezh2 wild-type and Ezh2 mutant H689A, and Dr. Eva Corey for LuCaP 35CR PDX. This work was supported, in part, by grants from Prostate Cancer Foundation grant (13YOUN007 to Q. Cao), U.S. Department of Defense grants (W81XWH-15-1-0639, W81XWH-17-1-0357, and W81XWH-19-1-0563 to Q. Cao), American Cancer Society grant (RSG-15-192-01 to Q. Cao), NIH/NCI grant (R01CA208257 to Q. Cao and Prostate SPORE P50CA180995 DRP), and Northwestern University Polsky Urologic Cancer Institute (to Q. Cao); and K. Chen was supported, in part, by grants from NIH/NHLBI (R01CA208257, HL100397, and HL099997) and Department of Defense (W81XWH-17-1-0357 and W81XWH-19-1-0563).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.