Cell-Free Biocatalysis for the Production of Platform Chemicals
- 1Department of Molecular Sciences, Macquarie University, Sydney, NSW, Australia
- 2Department of Molecular Medicine & Pathology, The University of Auckland, Auckland, New Zealand
- 3Biomolecular Discovery Research Centre, Macquarie University, Sydney, NSW, Australia
Genetically engineered host bacteria have an extensive history for the production of specific proteins including the synthesis of single enzymes for the modification of compounds produced for industrial purposes by biological or chemical processes. Such processes have been developed largely through the process of discovery. The ability to assemble multiple enzymes into synthetic pathways is a new development aided by the synthetic biology approach of constructing and assembling suitable enzymes into pathways that may or may not occur in Nature to provide a high-impact platform for bio-manufacturing of chemicals, biofuels and pharmaceuticals. Industry has depended on chemical catalysts because of the known constraints experienced frequently with biological catalysts but when high stereochemistry, mild synthesis conditions and environmentally friendly processes are significant, application of enzymes is preferred, especially in the case of drug synthesis where enantioselectivity is important. However, many whole cell production processes are beset by toxicity problems, metabolite competition, the production of side products, sub-optimal enzyme ratios and varying temperature optima. As a result, a cell-free biocatalysis allows the manipulation of substrate ratios, provision of regenerated cofactors and adjustment of high energy flux ratios that are difficult or impossible to control in whole cell synthesis. We discuss here the construction of cell free biocatalytic pathways as added free enzymes or multi-enzyme modules that may contain heterologous catalysts. We examine the status of applications leading to commercialisation, emphasizing the importance of economical cofactor regeneration. Nevertheless, problems remain, particularly protein post-translational modifications including glycosylation, phosphorylation, ubiquitination, acetylation, proteolysis, etc. The National Renewable Energy Laboratory and Pacific North West Laboratory have published lists of top value-added chemicals from biomass that include glucaric acid. There have been few reports on the combination of synthetic biology and cell-free protein synthesis to set up pathways for these valuable intermediate compounds. This observation is despite the existence of at least one large scale but specialized cell-free production of antibody conjugates. This review will provide a description of one successful attempt at the cell-free production of glucaric acid and will evaluate progress for other key intermediate and platform chemicals.
Introduction
The presentation of the European Commission’s Circular Economy Strategy and Strategic Plan in 2014 and Action Plan in 2015 have generated considerable discussion of the place, role and relationship of the Circular Bioeconomy as discussed in detail by Carus and Dammer (2018) and for a number of other individual and mainly agricultural industries such as forestry (DeBoer et al., 2020), ethanol production (Erickson, 2018), plastics (Blank et al., 2020); textiles (Aznar, 2019) and animals (Horton, 2019) along with many other examples. The circular bioeconomy requires absolute carbon efficiency and eventually, a move away from fossil feedstocks on which the chemical industry depends, largely crude oil and other fossil resources such as carbon and energy feedstocks. The contribution of biocatalysis to sustainable chemistry as part of the circular bioeconomy and the new ability to develop enzymatic transformations to fit new design parameters has been reviewed in depth by Sheldon and Woodley (2018). The energy industry may move to mixed carbon sources including conventional substrates (molasses and starch) and to non-conventional feedstocks such as CO2, methane, glycerol, agricultural and urban waste streams including CO2 and plastic wastes using biotechnological approaches (reviewed comprehensively by Clomburg et al., 2017). The question of “How can synthetic biology and cell free synthesis contribute to the aims of the global bioeconomy?” needs to be posed.
Enzyme technology plays an important role in synthetic biology and biomanufacturing (Bornscheuer et al., 2012; Reetz, 2013; Sheldon and Brady, 2018). Developments in a variety of tools have resulted in the classical concept of biocatalysis expanding from single enzyme reactions to the incorporation of multiple enzymes allowing the assembly of synthetic biocatalytic pathways (Bornscheuer, 2018; Poppe and Vértessy, 2018). The current trend for “green” synthesis of chemicals has resulted in an increased focus on biocatalysis as an alternative to traditional pathways for sustainable and environmentally responsible biomanufacturing (Swartz, 2006, 2012; Bornscheuer et al., 2012; France et al., 2017; Sperl and Sieber, 2018).
Enzymes are exceptional catalysts but many enzymes are tightly regulated and have not evolved naturally to function under most unnatural industrial conditions. Accordingly, industry has relied heavily on chemical catalysts although the application of enzymes is favored over conventional chemical catalysts (Keasling et al., 2012; Bornscheuer, 2016) particularly for biopharmaceuticals where there is a requirement for high optical purity with one specific enantiomeric form. As a result, single enzyme reactions have been gradually incorporated into industrial processes to complement chemical catalysis for enantioselective steps in a pathway. Several aspects of this challenging field were reviewed recently (Rudroff et al., 2018) and an expanded account of possibilities in the field is included in the review of Petroll et al. (2019). The interested reader is referred to a number of accounts aiming to solve specific problems and situations encountered in the transfer of enzymes to industrial reactions (Ye et al., 2012, 2013; DeSantis et al., 2003; Savile et al., 2010; Bornscheuer et al., 2012; Goldsmith and Tawfik, 2017; Gruber et al., 2017; Gumulya and Gillam, 2017; Wang et al., 2017; Risso et al., 2018; Kelwick et al., 2020).
Industrial biomanufacturing that incorporates single enzyme reactions use living cells (in vivo) which provide the advantage of enzymes being produced continuously and inside physical barriers like cell walls or internal compartments providing protection against enzyme degradation (Bornscheuer et al., 2012; France et al., 2017). In the case of multi-enzyme pathways in vivo the situation is more challenging and usually results in low yields because substrates and products may be channeled into different cellular metabolic pathways or display toxicity to the host cells (Guo et al., 2017). Also, the loss of recombinant plasmids that code for pathway enzymes in hosts impairs continuous cell-based biocatalysis (Friehs, 2004; Huang et al., 2012). Accordingly, multi-enzyme pathways are assembled with cell-free systems to allow the testing of an enzymatic pathway without the need for genetic alteration of the host organism and interference from other intracellular processes providing flexibility and control (Cherubini, 2010; Schmidt-Dannert and Lopez-Gallego, 2016; Taniguchi et al., 2017; Zhang et al., 2017; Wilding et al., 2018).
Cell-Free Enzyme Synthesis and Prototyping
Background
The use of multiple enzymes in a cell-free context has been available and practiced in the form of conventional cell-free synthesis (CFS) for over 40 years (Nirenberg and Matthaei, 1961; Devries and Zubay, 1967; Zubay, 1973). However, the rapid advance and development currently witnessed with synthetic biology has opened new possibilities for cell-free biocatalysis. Cell-free expression of proteins can be accomplished when all of the components necessary for RNA translation in vitro are present. The historical background of this field has been reviewed extensively and they will not be discussed here. However, the interested reader is referred to Swartz, 2006, 2012; Chong, 2014; Jewett et al., 2008; Yang et al., 2012; Whittaker, 2013: Rosenblum and Cooperman, 2014; Karim and Jewett, 2016; and Silverman et al., 2020 for further information. Although CFS was not considered as a suitable technology for large-scale protein production, Swartz and colleagues (Voloshin and Swartz, 2005) and Sutro Biopharma1 have reported high productivity with selected proteins at the hundred liters scale. The original CFS system was based on a crude cell lysate which was rapidly depleted of energy (e.g., ATP) and amino acids. In addition, the crude extract lysate was susceptible to macromolecular degradation by native proteases. The Swartz group refined the lysate system to address the issues of toxicity, transport limitation, cell membrane effects and also supplied alternative energy sources to overcome the problems encountered by the original system (summarised in Swartz, 2006, 2012). Another system was developed by Shimizu et al. (2005) using 32 purified factors for all aspects of protein synthesis allowing the production of enzymatically active proteins. This system is now available as PURExpress (New England Biolabs) and although it is suitable for small scale protein production the cost of the components prohibit its application for large-scale protein production.
We have concentrated on cell free protein synthesis. We do not review other high value compounds and substrates such as hydrogen and bioelectricity. The interested reader will find accounts of applications and potential commercialisation prospects in the account from the US Department of Energy, Office of Energy Efficiency & Renewable Energy. (2018) and Shi et al. (2018), as well as many other papers and books on these topics.
Advantages of Cell-Free Enzyme Synthesis
Recently, the scope and principles of CFS have been expanded by Karim and Jewett (2016) and comprehensively reviewed by Silverman et al. (2020). Karim and Jewett suggested a framework for biosynthetic pathway development based on CFS-driven metabolic engineering. The framework enabled the optimisation of a pathway comprised of 17 enzymes by using heterologous enzymes from Clostridium beijinkerii, C. acetobutylicum and Treponema denticola. These enzymes were used to supplement the native Escherichia coli enzymes for the production of n-butanol from glucose (Karim and Jewett, 2016). This study demonstrated that the cell-free protein expression simulated the in vivo system and its potential for prototyping biocatalytic pathways faster and easier than using metabolic engineering. This mode of development has been emphasized in the recent description of the cell-free styrene biosynthesis platform that brought about the conversion of L-phenylalanine to styrene using carefully titrated phenylalanine ammonia lyase to produce the intermediate trans-cinnamic acid, which was treated with ferulic acid decarboxylase to give styrene along with ammonia and carbon dioxide as side products (Grubbe et al., 2020).
Examples of synthetic cell-free pathways based on enzymes or culture lysates are given in Table 1 of this review. Most of these pathways use simple and renewable substrates like glucose or starch and have shown to reach theoretical conversion yields of nearly 100%. Several of the intermediates or products in these pathways are central carbon metabolites (e.g., pyruvate) and most of them would have been channeled into the cell’s own metabolism if the reaction was performed in vivo. Moreover, often these reactions are performed at temperatures higher than 37°C which may be limiting in vivo while many products like terpenes (Korman et al., 2017) and isobutanol (Guterl et al., 2012) are produced at the concentrations that would be toxic to cells.
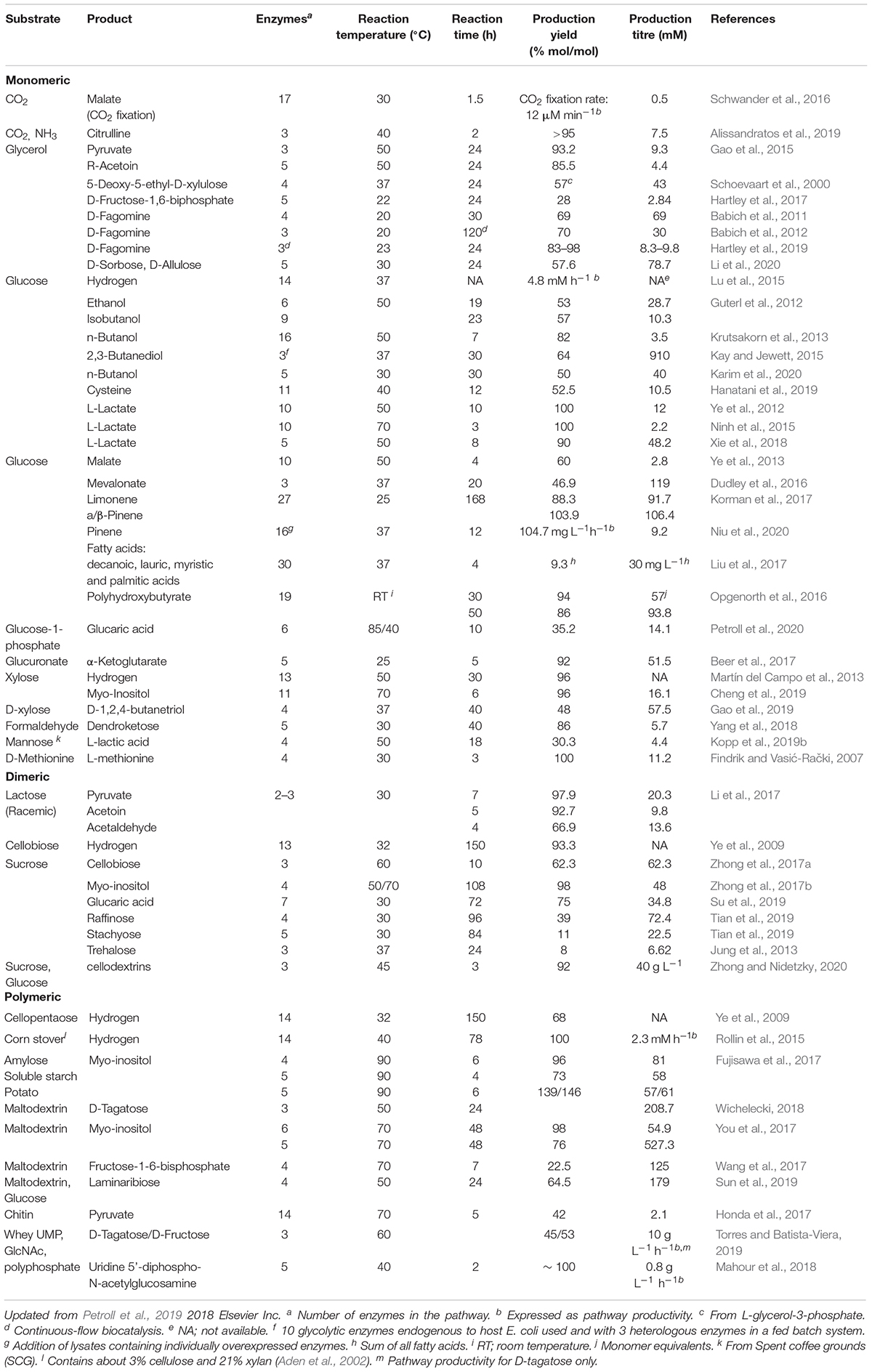
Table 1. Cell-free enzyme pathways reported for the production of biofuels or high value chemicals from different substrates.
Cell-free biocatalysis has become an intrinsic part of the synthetic biology revolution and in this context it has been referred to lately as cell-free synthetic biology. However, it is important to emphasize that assembling multi-enzyme functional biocatalytic pathways using enzymes from different organisms is not a simple exercise and requires a deep understanding of all parameters that might affect enzyme activity and stability under different operational conditions.
Prototyping
Several research groups over the last decade have taken advantage of the use of CFS to overcome barriers such as the cell wall and the fact that the usual microbial systems carry on with growth as expressed by a number of metabolic pathways in addition to the pathway of interest. The development of the field and the advantages the prototyping process offers has been reviewed in depth recently (Silverman et al., 2020). Cell-free systems have long been used as single enzyme approaches for the improvement of yields of particular biochemicals. Recently, there has been emphasis on multi-enzyme complexes in cascades in the development of pathway engineering and it is clear that the rapid rise of synthetic biology owes much to the controllable cell-free environment, the ability to direct substrates toward the desired product and the facility with which superior combinations of often heterologous enzymes can be assembled. These issues have been reviewed in detail recently and the reader is referred to Jewett et al. (2008); Carlson et al. (2012), Gan and Jewett (2014), Hodgman and Jewett (2013), Karim and Jewett (2016, 2018), Kelwick et al. (2016), Moore et al. (2017b), and Silverman et al. (2020). Further descriptions of pathway developments (for example, Dudley et al., 2019, 2020 with limonene and isoprenoid synthesis). There is the possibility of the synthesis of products that have been prototyped with freeze-dried protein synthesis components (Pardee et al., 2016; Pardee, 2018) as well as opportunities offered for the design and engineering of systems from the bottom up in prototyping procedures as part of the design-build-test cycle. Cell-free synthesis prototyping has been confined largely to E. coli as a model organism and takes two forms. The PURE system relies on purified cellular components (108 in total, Shimizu et al., 2001) and is available commercially (New England Biolabs) but its high cost has prevented scale up. Its main use is in small scale prototyping (Moore et al., 2017b). A less expensive route is to employ crude cell lysates where the results are scalable into high volume fermentations (Zawada et al., 2011; Cai et al., 2015) coupled with inexpensive energy sources and an awareness of the problem of diversion of at least some of the target product into other reactions. The question of the applicability of CFS to the production of certain typical products has been roughly evaluated by Silverman et al. (2020) and shown to be related to the cost price of the bulk chemical produced by whole cell fermentation on one hand and the production of bio-pharmaceuticals and vaccines by CFS on the other.
A recent preprint from the Jewett group (Krüger et al., 2020) describes the use of prototyping for optimisation of protein yield of a non-traditional but industrially relevant Clostridial host as judged by luciferase bioluminescence. The cell free system allowed rapid examination of a number of metabolites, pathways and energy regeneration systems which were shown to vary substantially from the standard E. coli conditions. They also showed how the system could be used for increasing expression of protein synthesis for the selected gene in C. autoethanogenum, with the possibility of adaptation and optimisation of the prototyping system for improvement of yields by solventogenic and cellulolytic Clostridia.
Applications and Scale Up
While the metabolic engineering field has traditionally centered on cell-based production, cell-free systems in the form of multi-enzyme biocatalysts have developed as an alternative system in which a mixture of enzymes and cofactors support the conversion of a substrate into a product. Construction of pathways in a cell-free environment allows for more flexibility and engineering freedom and cell-free pathway engineering is developing toward a truly synthetic approach for metabolic pathway design. In the design space, the only limitations are set by stoichiometry, thermodynamics and influence of the intermediates and cofactors brought into or produced within the system. Therefore, cell-free approaches enable the implementation and combination of pathways found in nature, pathways with novel enzymes and pathways with novel chemistries.
Cell-free synthesis has served as a platform technology for a number of biotechnology and synthetic biology procedures and the literature is replete with reviews and papers describing its utilization (for example, Rosenblum and Cooperman, 2014; Smith et al., 2014; Khambhti et al., 2019; Rolf et al., 2019; Tinafar et al., 2019). This phenomenon may be linked to its ability to synthesize proteins that are difficult to produce with whole cells but also its uses in rapid pathway prototyping, allowing accelerated selection of advantageous mutants (comprehensively reviewed by Karim and Jewett, 2016, 2018). Cell-free synthesis has been central to diagnostic assays based on CRISPR-cas9 and its adaptations for field use in epidemic situations (see Marshall et al., 2018; Pardee, 2018) as well as protein microarrays, microchips and miniaturization (summarized in Rolf et al., 2019; Ayoubi-Joshaghani et al., 2020). For the interested reader, a comprehensive review of the field is provided by Swartz (2018) but in general, most applications have been at the level of the bench and the commercialisation of the process has lagged behind the academic science.
Substrates for Biomass Transformation
Enzymes and cofactors are mixed together in cell-free biocatalysis to achieve the conversion of a substrate into a final product (Sheldon and Woodley, 2018; Wilding et al., 2018). In an industrial context, several challenges must be met by cell-free biocatalysis in order to be competitive with established chemical practices. They include robustness and efficiency of the biocatalytic system, self-sufficiency of cofactor requirements and resistance to inhibitory side products (Schmidt-Dannert and Lopez-Gallego, 2016). Coupling the activities of several enzymes and cofactors into cascade reactions have been implemented to address the necessary demands for the potential industrial application of cell-free biocatalysts (Figure 1). Cell-free biocatalysis using multi-enzyme cascades have been designed in the last decade to assemble complete central metabolic pathways e.g., glycolysis (Krutsakorn et al., 2013; Korman et al., 2017), TCA cycle (Sokic-Lazic and Minteer, 2009) and fatty acid biosynthesis (Yu et al., 2011). Cell-free biocatalytic systems have been used in the assembly of natural and synthetic pathways for the production of relevant chemicals including alcohols (Guterl et al., 2012), polymers (Opgenorth et al., 2016), terpenes (Korman et al., 2017) and sugar fermentation and oxidation (Guterl et al., 2012; Xu and Minteer, 2012; Opgenorth et al., 2016; Zhu and Zhang, 2017).
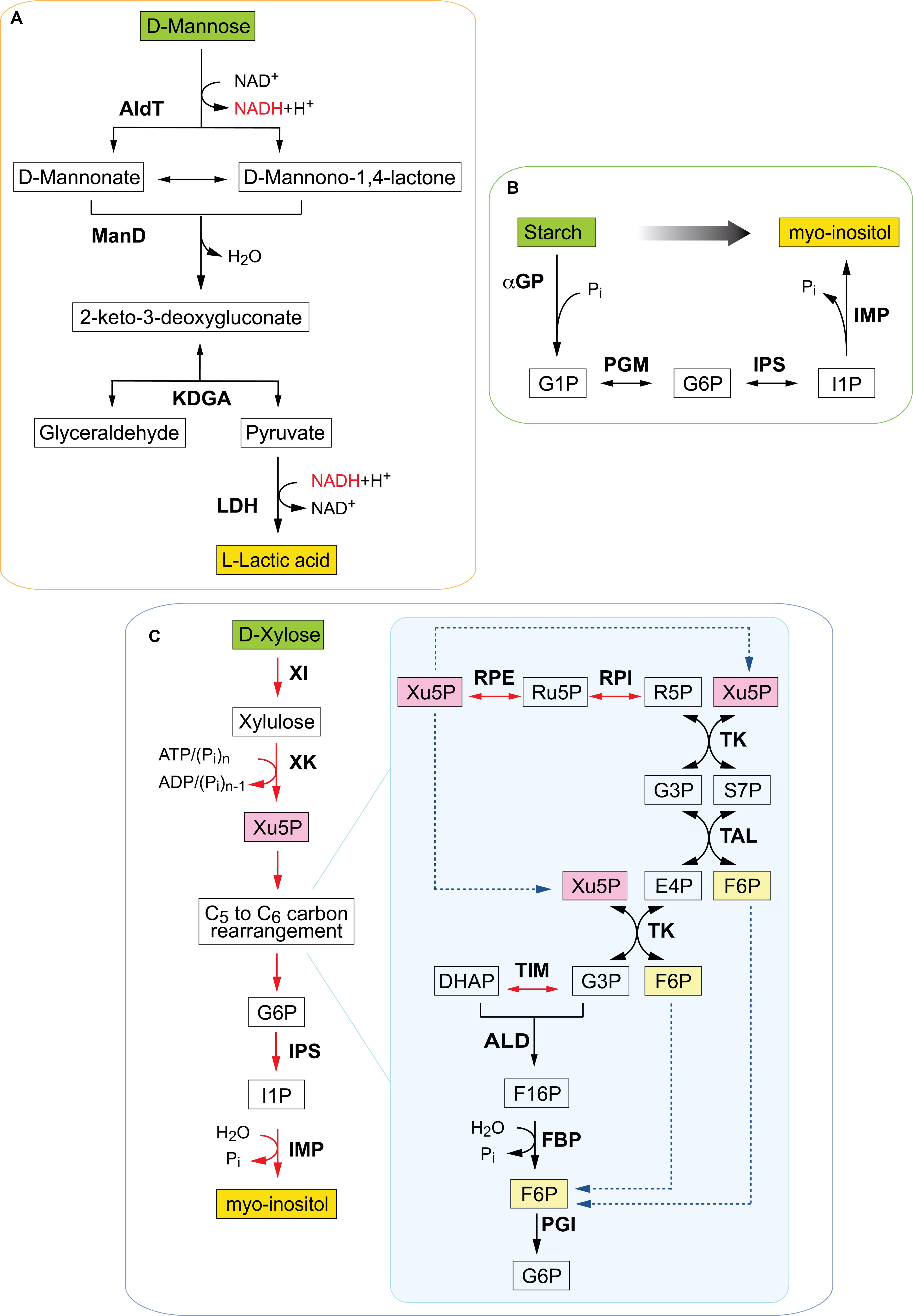
Figure 1. Schematic illustration of selected in vitro synthetic pathways assembled for different monomeric and polymeric substrates. (A) Synthetic enzymatic pathway for cell-free conversion of mannose obtained from spent coffee grounds into lactic acid. Enzymes are in bold, reduced cofactor in red. AldT, aldohexose dehydrogenase; ManD, mannonate dehydratase; KDGA, 2-keto-3 deoxygluconate aldolase; LDH, lactate dehydrogenase; G3P, glyceraldehyde-3-phosphate; DHAP, dihydroxyacetone phosphate. Adapted from Kopp et al. (2019b). (B) Non-fermentative cofactor-free enzymatic pathway for the inositol synthesis from starch. Enzymes include αGP, α-glucan phosphorylase; PGM, phosphoglucomutase; IPS, inositol-1-phosphate synthase; IMP, inositol monophosphatase. Chemical compounds are G1P, glucose 1-phosphate; G6P, glucose 6 phosphate; I1P, inositol 1-phosphate; Pi, inorganic phosphate; Glc, glucose; (Glc)n, starch or maltodextrin with the number of degree of polymerization of n; ATP, adenosine triphosphate; ADP, adenosine diphosphate. Adapted from You et al. (2017). (C) Synthetic enzymatic pathway for the synthesis of myo-inositol from D-xylose. Enzymes are XI, xylose isomerase; XK, xylulokinase; RPE, ribulose 5-phosphate epimerase; RPI, ribose 5-phosphate isomerase; TK, transketolase; TAL, transaldolase; TIM, triose phosphate isomerase; ALD, fructose- bisphosphate aldolase; FBP, fructose 1,6-bisphosphatase; PGI, phosphoglucose isomerase; IPS, inositol 1-phosphate synthase; IMP, inositol monophosphatase. Key metabolites include Xu5P, xylulose 5-phosphate; G6P, glucose 6-phosphate; Ru5P, ribulose 5-phosphate; R5P, ribose 5-phosphate; S7P, sedoheptulose 7-phosphate; G3P, glyceraldehyde 3-phosphate; E4P, erythrose 4-phosphate; DHAP, dihydroxyacetone phosphate; F16P, fructose 1,6 bisphosphate; F6P, fructose 6-phosphate; I1P, inositol 1-phosphate. Adapted from Cheng et al. (2019).
So far, multi-enzyme biocatalysis has relied heavily on substrates obtained from refinery (glucose) or fermentation processes (glucose-6-phosphate) (Woodward et al., 2000; Guterl et al., 2012; Ye et al., 2012; Krutsakorn et al., 2013; Dudley et al., 2016; Liu et al., 2017; Zhu and Zhang, 2017). The concept of a circular economy is based on waste minimisation, valorisation and recycling (Imbert, 2017) and as such, the utilization of waste as substrate is central for developing sustainable biomanufacturing processes. Demand from national and international organizations as well as increasing public environmental and socio-economic concerns have shifted the attention to the use of renewable waste as a source of substrates for biocatalysis (Koutinas et al., 2014; Bilal and Iqbal, 2019). Diverse waste streams which include agricultural, industrial and municipal wastes are rich in carbohydrates, proteins and lipids, making them a potential feedstock for enzymatic processes to produce high value bioproducts. Biopolymers like cellulose, hemicellulose, lignin and starch have been recognized particularly as substrates for biofuel production (Pleissner and Lin, 2013). However, recent studies have shown that the conversion of biomass waste into bulk chemicals is 3.5 times less expensive than converting it into biofuels (Tuck et al., 2012).
An estimated 140 billion m3 tons of biomass wastes are generated annually from agriculture (United Nations Environment Programme [UNEP], 2015). While the average agricultural biomass contains approximately 50–60% cellulose/hemicellulose and 15–25% lignin (Kenney et al., 2013) less attention has been given to other natural sources of polysaccharides including marine polysaccharides and plant gums (Mukherjee and Gupta, 2015). An estimated 1.3 billion tons of food are wasted globally each year (Food and Agriculture Organization [FAO], 2011). UNEP in partnership with the FAO are focused on reducing food waste by 50 per cent by 2030 (United Nations Environment Programme [UNEP], 2018). Food processing results in the production of wastes in the form of liquids, solids, or semi-solids. Many food industries utilize large quantities of water in sanitation, cleaning and cooking processes and generate effluents that contain diluted organic compounds, nitrogen and fats. Examples include potato-processing wastewater, fruit residue sludges, whey from dairy manufacturing, bakery and brewery industry effluents. On the other hand, solid food wastes are rich in polysaccharides (e.g., lignin, cellulose, starch) and monosaccharides (e.g., glucose and fructose). Examples include potato and tomato wastes, wastes from bread production, apple and grape pomace and soybean curd residue (Tuck et al., 2012; Hegde et al., 2018; Bilal and Iqbal, 2019). Some examples of potential renewable substrates currently used for cell-free multi-enzymatic biocatalytic systems are presented below and summarized in Table 1.
The corn milling process produces corn steep water as by-product. It is rich in proteins and contains simple and complex carbohydrates which can be converted further to racemic lactate through Lactobacillus-aided fermentation (Hull et al., 1996). However, many lactate-based industries demand optically pure lactate. Several cell-free multi-enzyme pathways were assembled by Li et al. (2017) using D-lactate oxidase, L-lactate oxidase, pyruvate decarboxylase and catalase for the conversion of racemic lactate from corn steep water into acetaldehyde, pyruvate and acetoin. The pathways were designed to include stereo-selective oxidases to produce optically pure L- or D-lactate.
In an earlier study, Rollin et al. (2015) demonstrated for the first time the production of hydrogen directly from pre-treated plant biomass using cell-free biocatalysis. Corn stover was converted to xylose and glucose using acid hydrolysis and enzymatic degradation and the sugars were simultaneously converted to hydrogen using a cell-free synthetic pathway assembled with more than 10 purified enzymes. A combination of kinetic modeling, improved enzyme loading, increased substrate concentration and reaction temperature resulted in a 67-fold increase in productivity with a final production of 54 mmol H2 L−1 h−1.
Chitin is the second most abundant organic compound on earth and is a polymer made of β-1,4-linked-N-acetylglucosamine subunits. It is found primarily in the exoskeleton of crustaceans and fungal cell walls (Gooday, 1990). Worldwide, more than 6 million tons of waste containing chitin is produced annually mainly from the crab, shrimp and lobster industries (Yan and Chen, 2015). To address the challenge of using a single bioreactor system that combined enzymatic saccharification and microbial fermentation at different optimum temperatures, Honda et al. (2017) designed a one-pot cell-free multi-enzyme pathway for the conversion of colloidal chitin into pyruvate. The pathway incorporated 12 thermophilic enzymes which were produced heterologously in E. coli and obtained after a heat treatment of the cell extract. The system combined a chitin degradation module and a cofactor-recycling module and produced 0.62 mM pyruvate in 5 h from 0.5 mg mL–1 colloidal chitin at 70°C. However, the long-term bioconversion was affected by the use of semi-purified enzyme preparations which resulted in ATP degradation by the host enzyme(s).
Coffee is one of the world’s largest traded commodities with an annual production of almost 10 billion kg beans (International Coffee Organization, 2019). However, only 30% of the coffee grounds are solubilised during coffee preparation, creating a waste stream that primarily ends up in landfills (Fernandes et al., 2017). Spent coffee grounds (SCG) are the primary end product of industrial coffee production and beverage preparation and are rich in polysaccharides, lipids and proteins (Massaya et al., 2019). In the context of a circular economy, SCG are an under-utilized industrial waste resource with potential of becoming an ideal feedstock for enzymatic valorisation in the production of fine chemicals. Reutilisation of SCG waste has been reported for the production of biofuels (Caetano et al., 2012; Vardon et al., 2013), as a source of bioactive secondary metabolites (Ramalakshmi et al., 2009), immunostimulatory oligosaccharides (Takao et al., 2006) and for the extraction of phenolic compounds (Zuorro and Lavecchia, 2012). SCG is made up of approximately 50% carbohydrates and is rich in galactomannan, a polysaccharide containing a mannose backbone with galactose side groups. However, only a few studies have been undertaken specifically to utilize this component of SCG (Campos-Vega et al., 2015). One example is the recent work reported by Kopp et al. (2019a) who assembled a functional non-phosphorylative synthetic pathway to convert mannose obtained from SCG into lactic acid (Figure 1A). The cell-free multi-enzyme pathway comprised three thermostable enzymes from the thermoacidophilic archaeon Thermoplasma acidophilum, including a newly identified mannonate dehydratase and a 2-keto-3-deoxygluconate aldolase (Kopp et al., 2019b). The last enzyme of the pathway, a lactate dehydrogenase from Bacillus stearothermophilus, catalyzed the conversion of pyruvate into lactic acid. The pathway assembled represents the shortest known enzymatic conversion of mannose into lactic acid and was able to produce 4.4 mM lactic acid from 14.5 mM SCG-derived mannose.
Glycerol is a versatile non-toxic, biodegradable and recyclable “green solvent” which is the main coproduct in the biodiesel process. Global biodiesel production has grown steadily in recent years with more than 30 million m3 produced in 2015 (REN21,2017). During the biodiesel process the transesterification reaction produces biodiesel and glycerol, with glycerol comprising 10% of the total biodiesel generated (Johnson and Taconi, 2007). In 2011, only 40% of the 5.1 million tons of the glycerol produced worldwide was used and with a global production expected to surpass 7.6 million tons by 2020, the surplus is anticipated to increase dramatically (Anuar and Zuhairi, 2016). Consequently, currently there is an urgent need for alternative applications for the use of surplus glycerol. Accordingly, there is growing interest in the use of glycerol as substrate for biocatalysis and its conversion into value-added chemicals. One of the first cell-free enzymatic cascades for the conversion of glycerol into pyruvate was reported by Gao et al. (2015). The simplified synthetic pathway consisted of only three enzymes (alditol oxidase, dihydroxy acid dehydratase, and catalase) without the need for NAD+/NADH as cofactors. However, this initial pathway was not suitable for the production of chemicals with higher degrees of reduction (e.g., lactate, alanine, 2,3-butanediol and ethanol). An improved cell-free multi-enzyme pathway was described recently for the production of L-lactate from glycerol using four thermostable enzymes (alditol oxidase, dihydroxy acid dehydratase, catalase and L-lactate dehydrogenase) coupled to an NADH regeneration system using a formate dehydrogenase. The modified system produced 34.4 mM L-lactate from 50 g/L glycerol after 72 h at 50°C (Li et al., 2018).
Several enzymatic cascades have been reported for the synthesis of non-natural carbohydrates from glycerol. For example, Schoevaart et al. (2000) assembled an alternative four enzyme pathway based on phosphorylation of glycerol with a phytase enzyme. The one-pot enzyme reaction combined four different enzymes; a phytase, glycerol phosphate oxidase, catalase and fructose-1,6-biphosphate aldolase to produce 5-deoxy-5-ethyl-D-xylulose from glycerol. Starting from glycerol and pyrophosphate, a 57% yield of 5-deoxy-5-ethyl-D-xylulose was obtained from l-glycerol-3-phosphate. In another study, the conversion of glycerol to dihydroxyacetone phosphate (DHAP) was reported with a one-pot four-enzyme cascade composed of glycerol kinase (Thermococcus kodakarensis), acetate kinase (Mycobacterium smegmatis), glycerophosphate oxidase (Mycoplasma gallisepticum) and catalase (Micrococcus lysodeikticus) (Hartley et al., 2017). The cascade achieved 88% conversion of glycerol to DHAP in 1 h at 22°C. The pathway was coupled with a DHAP-dependent fructose-1,6-biphosphate aldolase enzyme from Staphylococcus carnosus to produce the rare chiral sugars D-fructose-1,6-biphosphate (3S, 4R) and 3,4-dihydroxyhexulose phosphate (3S, 4R). D-fagomine is a naturally occurring iminocyclitol (iminosugar) found in plants which has an antihyperglycemic effect (Asano et al., 1994). Recently, Hartley et al. (2019) implemented a novel approach to convert glycerol into D-fagomine precursor using single-molecule multi-enzyme biocatalysts that retain and recycle their cofactors (Figure 2). A three-step cascade initially was used to phosphorylate glycerol which was then oxidized to form dihydroxyacetone phosphate. In the final step, D-fagomine was produced via an aldolase-dependent reaction. This three-step cascade was integrated into a continuous-flow reactor system that produced up to 70 g L–1 h–1 g–1 D-fagomine and thus displayed higher yields compared to previously reported chemical syntheses (Castillo et al., 2006).
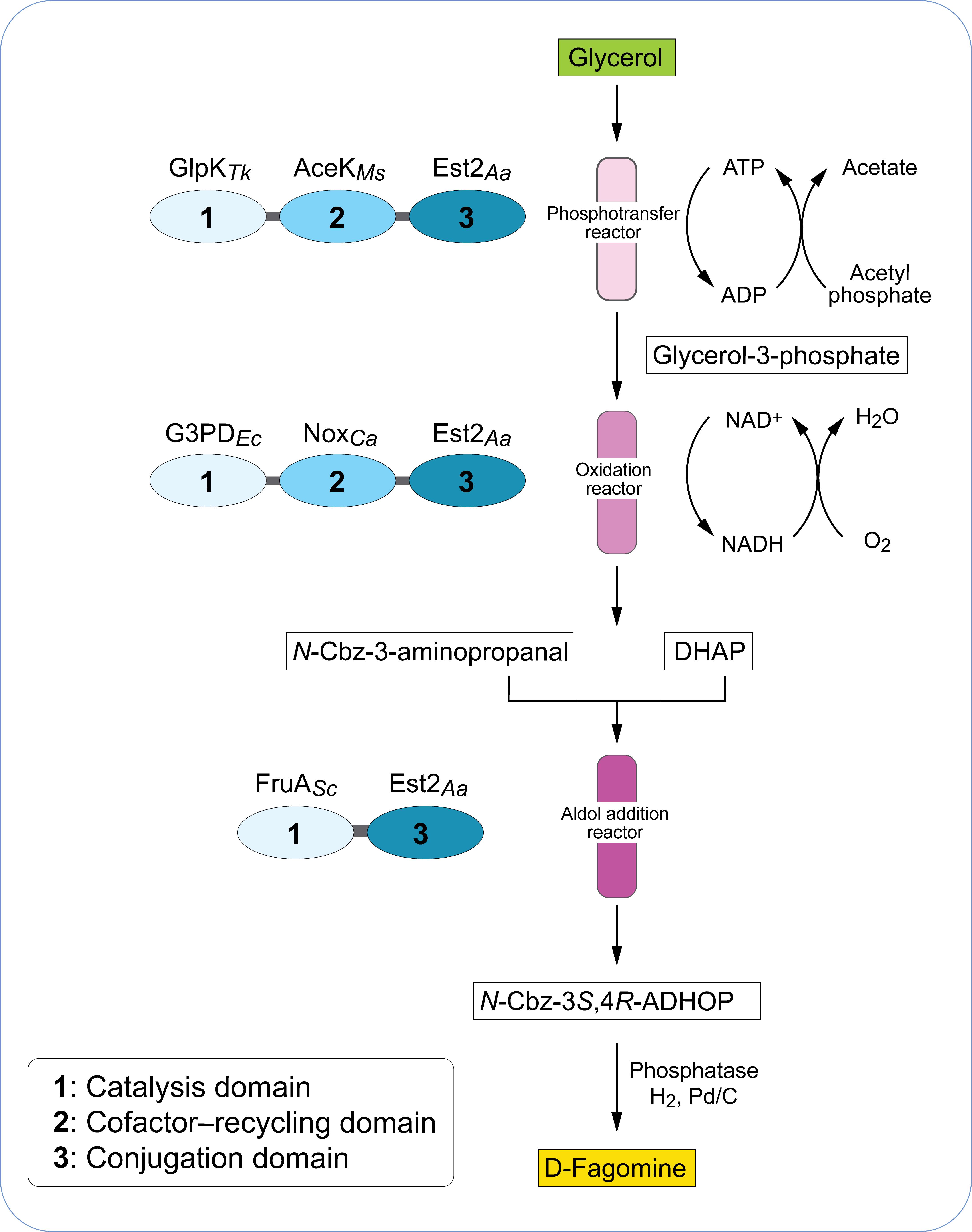
Figure 2. Biocatalyst and reactor design of enzymes that retain and regenerate their cofactors. Each biocatalyst consists of three protein modules: a cofactor-dependent catalytic enzyme module (1), a cofactor-recycling module (2) and a conjugation module (3). Modules are connected by short amino acid spacer regions (gray). The modules are combined in a three-step conversion of glycerol to a D-fagomine precursor using a continuous-flow reactor. DHAP, dihydroxyacetone phosphate; N-Cbz-3S,4R-ADHOP, N-Cbz-3S,4R-amino-3,4-dihydroxy-2-oxyhexyl phosphate. GlpKTk, glycerol kinase from Thermococcus kodakarensis; AceKMs, acetate kinase from Mycobacterium smegmatis, Est2Aa, esterase E2 from Alicyclobacillus acidocaldarius; G3PDEc, glycerol-3-phosphate dehydrogenase from Escherichia coli; NoxCa, water-forming NADH oxidase from Clostridium aminovalericum; FruASc, fructose aldolase from Staphylococcus carnosus. Adapted from Hartley et al. (2019).
Plant biomass degradation has been reviewed at a top down level by Schillberg et al. (2019) who address broadly the problems associated with the production of proteins but conclude that some plant-derived recombinant proteins are competitive in niche areas. Wu et al. (2017) described the isolation of a potential thermophilic cellodextrin phosphorylase that was able to use long chain cellodextrins for the synthesis of oligosaccharides. The potential for the production of pyruvate from glucose was discussed as part of a general approach to the degradation of biomass with emphasis on the growth conditions used for the generation of the cell free extracts (Garcia et al., 2018).
Multi-enzyme biocatalytic systems show great potential for effectively catalyzing value-added bioconversion processes. At the center of the biobased economy movement is the substitution of oil-derived products with products obtained from renewable and socially responsible resources such as plant-derived carbohydrates, lipids from algae or waste streams. The examples provided above are only a limited sample of waste streams and industrial by-products that are available as potential substrates for cell-free multi-enzyme based biomanufacturing. Any effective utilization of these substrates will need consideration of their chemical complexity and the ability to direct their degradation products into a multi-enzyme system without compromising enzyme activities and production costs.
Cofactor Regeneration Systems
Many enzymes rely on cofactors for their catalytic activity (Zhao and van der Donk, 2003). Cofactors are low molecular weight organic or inorganic compounds that play a major role in cell-free biocatalysis where reactions rely on the supply of exogenous cofactors which are expensive supplements in industrial biocatalysis. For example, with the approximate cost of NADH at $260 per gram and for cell-free biocatalysis to be economically attractive, cofactors must be regenerated and recycled (Britton et al., 2018). Accordingly, cofactor cost and recycling have hampered the implementation of cell-free multi-enzyme biocatalysis at the industrial level and many efforts have been made to avoid their use (Welch and Scopes, 1985; Opgenorth et al., 2014; Nowak et al., 2015; Honda et al., 2016; Beer et al., 2017). The interested reader is referred to Zhao and van der Donk (2003) and Andexer and Richter (2015) for a more comprehensive review on cofactor regeneration and ATP regeneration systems, respectively. Expensive cofactors also can be replaced using synthetic NAD-based biomimetics developed to retain the enzyme-recognition moieties and which can be engineered to be more stable than that their natural counterparts (Nowak et al., 2015, 2017). A more expanded discussion on the application of synthetic NAD-based biomimetics is provided by Paul and Hollmann (2016).
Cell-free cofactor regeneration and recycling can be achieved by integrating auxiliary enzymatic reactions which regenerate the spent cofactor using an inexpensive substrate cofactor to reduce dependency and costs. For example, NAD+ can be recovered by adding a water-forming NADH oxidase that oxidizes NADH to NAD+ (Geueke et al., 2003; Gao et al., 2012; Nowak et al., 2015; Beer et al., 2017). Similarly, NADH can be recovered from NAD+ and from less expensive substrates (e.g., formate or glucose) using a formate or glucose dehydrogenase (Van der Donk and Zhao, 2003). For ATP regeneration, the incorporation of polyphosphate kinases, which accept inexpensive polyphosphate, can be used to synthesize ATP from ADP or AMP in ATP-dependent reactions (Nocek et al., 2008; Schwander et al., 2016).
It is highly desirable to use more robust thermostable enzymes in many applications. In such conditions biocatalysis is usually performed at temperatures above 50°C at which heat-labile cofactors including NAD(P)H and ATP tend to degrade rapidly. In cell-free systems, the loss of heat-labile cofactors at these higher temperatures must be overcome by continuous supply of exogenous cofactor resulting in a more costly process (Ye et al., 2012; Huang et al., 2016). Several alternative strategies have been developed to address this challenge. In one example, a chimaeric version of the Embden-Meyerhof-Parnas pathway was developed using exclusively thermophilic enzymes from bacteria and archaea. In this non-ATP forming pathway, ATP-balanced conversion of glucose to lactose was achieved by replacing the enzyme pair glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate kinase by a non-phosphorylative glyceraldehyde-3-phosphate (Ye et al., 2012). A multi-enzyme, cell-free NAD salvaging pathway was reported which was composed of 8 thermostable enzymes produced heterologously in E. coli. The heat-treated crude extracts were used directly as the enzyme source and were capable of sustaining a constant NAD+ concentration for 15 h at 60°C (Honda et al., 2016).
Oxidative non-phosphorylative pathways also have been used to circumvent cofactor dependence in cell-free multi-enzyme biocatalysis. For example, Guterl and co-workers reported the production of ethanol and isobutanol from glucose by assembling a combination of the archaeal non-phosphorylative Entner-Doudoroff (np-ED) pathway and the isobutanol and ethanol pathways (Guterl et al. 2012). In the initial step glucose was oxidised to pyruvate via a cofactor-balanced pathway which relied on enzymes of the archaeal np-ED. In the second step, pyruvate was converted to either isobutanol or ethanol depending on the enzyme selection. While a conventional glycolytic pathway requires 13 enzymes, as well as NADH and ATP for the conversion of glucose to ethanol (Welch and Scopes, 1985), the modified oxidative non-phosphorylative pathway required only 6 enzymes and a single cofactor. Similarly, Xie et al. (2018) used a cofactor (NADH)-balanced cell-free pathway without ATP and only 5 thermophilic enzymes to produce lactate from glucose via pyruvate, achieving a product yield of ∼90%. Oxidative non-phosphorylative pathways also are available for sugars other than glucose (e.g., mannose, arabinose, galactose, and xylose) that are abundant in biomass and thus provide more opportunities to assemble cell-free multi-enzyme pathways with minimum cofactor requirements. Recently, a cell-free reaction cascade (Figure 1A) for the conversion of mannose into lactic acid was reported based on a so far unknown oxidative mannose metabolism from Thermoplasma acidophilum (Kopp et al., 2019a, b). The pathway was assembled using only 4 thermostable enzymes to achieve the bioconversion and was free of phosphorylated intermediates and cofactors.
Other strategies to overcome cofactor limitation and recycling include the design of self-sufficient heterogenous biocatalysts by co-immobilizing enzymes and cofactors onto solid phases in which enzyme and cofactor remain catalytically active and available for several reaction cycles. However, systems like these have shown reduced enzymatic activity toward the immobilized cofactors, limited recycling of immobilized enzymes and cofactors, displayed low turnover numbers of immobilized cofactors and low scalability of the overall system (El-Zahab et al., 2008; Fu et al., 2014; Beauchamp and Vieille, 2015; Ji et al., 2015). To overcome these limitations, Velasco-Lozano and co-workers designed self-sufficient heterogeneous biocatalysts with enzymes and cofactors within the same porous microbead (Velasco-Lozano et al., 2017). The enzymes were irreversibly bound to the polyethyleneimine(PEI)-coated agarose microbeads while the negatively charged cofactors were reversibly absorbed onto the positively charged PEI attached to the microbead surfaces. As a result of the dynamic adsorption via ion-exchange interactions to the solid surface or porous space, the cofactors are in a continuous association/dissociation equilibrium, with an exchange between enzyme active sites and the microbead surfaces, resulting in a self-regenerating system. This technique was used to design a self-sufficient heterogenous biocatalyst which consisted of two dehydrogenases co-immobilized with NAD+ for the symmetric reduction of ketones with 100% conversion yield and without the need of adding exogenous cofactor. The application of self-sufficient heterogeneous biocatalysts was expanded to the successful synthesis of chiral alcohols using a NADPH-dependent ketoreductase co-immobilized with NADPH to catalyze asymmetric reductions without added exogenous cofactor (Benítez-Mateos et al., 2017) and for the co-immobilization of ω-transaminases and pyridoxal 5′-phosphate for the enantioselective deamination of model amines (Benítez-Mateos et al., 2018).
In another recent elegant approach, Hartley et al. (2019) designed a single molecule biocatalyst which was composed of three different modules (Figure 2); a catalytic module (drives synthetic reaction), a cofactor-recycling module (regenerates cofactor) and an immobilization module (mediates conjugation to surface). In this strategy, all three modules were expressed as a single recombinant protein in E. coli with each module separated by a short amino acid spacer. This system was used in a three-step continuous flow-reactor for the production of D-fagomine precursor. In the reaction, glycerol was converted via an initial phosphorylation and then an oxidation reaction to dihydroxyacetone phosphate which served as substrate in a subsequent cofactor-independent aldolase-catalyzed aldol addition reaction. The combination of this single-molecule multi-enzyme biocatalyst and the continuous flow-reactor system resulted in up to 70 g L–1h–1g-1 of D-fagomine precursor.
In a circular economy, utilization of organic wastes as substrates is central for developing sustainable biomanufacturing processes. Similarly, avoidance of unfavorable cofactor dependency, which might result in more costly processes, is critical for the competitiveness of cell-free biocatalysis against established practices.
Cell-Free Enzyme Pathways
Recent examples of cell-free enzyme pathways which utilize from 3 to 30 enzymes either as cell lysates or purified enzymes are presented in Table 1. In most of the pathways, the product titres approach their maximum theoretical yields but there are some drawbacks such as intermediates that are diverted toward other cellular metabolites, process temperature and toxicity. We have focussed on glucaric acid biosynthesis but the Table illustrates how diverse this research area has become in a relatively short time. It shows also how sophisticated some of the pathways have become in the development of complex enzyme cascades. This outcome is reflected in many areas where research has been aimed at greener solutions to energy production, plastic pollution, upgrading of waste materials and replacement of petroleum-based products by biologically based ones of a value-added nature. One trend that is apparent has been the use of thermophilic enzymes in many pathways to take advantage of the robust nature of these proteins. We review below some of the many publications on relevant cell-free approaches to some of the problems indicated and acknowledge that we have not included many papers that are also significant.
Production of glucaric acid has attracted attention since being named as a top value-added chemical from biomass by the United States Department of Energy as it has shown value as a therapeutic agent and has a role in the chemical industry as an intermediate in slow-release fertilizers, biodegradable polymers and detergents. Most glucaric acid is produced by nitric acid oxidation of glucose and attempts at whole cell fermentative production by E. coli and yeast have not been economically viable (Moon et al., 2009; Gupta et al., 2016). A cell-free synthetic pathway was described by Su et al. (2019) that used seven enzymes in a one-pot reaction at a moderate temperature in the production of glucaric acid from 50 mM sucrose, producing 35 mM glucaric acid in 70 h (75% molar yield) (Figure 3A). The kinetic parameters of the rate- limiting enzyme, myo-inositol oxygenase (MIOX) were compared from various biological sources and the one from Arabidopsis thaliana was reported to have the highest specific activity. They used the addition of 10% NaOH in fed-batch mode to maintain the stability of enzymes which could be further improved via immobilization. Glucaric acid synthesis decreased when concentrations of over 50 mM sucrose were used or after 24 h when additional MIOX supplementation was required. The pathway used NOX (NADH oxidase, EC 1.6.3.1) enzyme for cofactor regeneration.
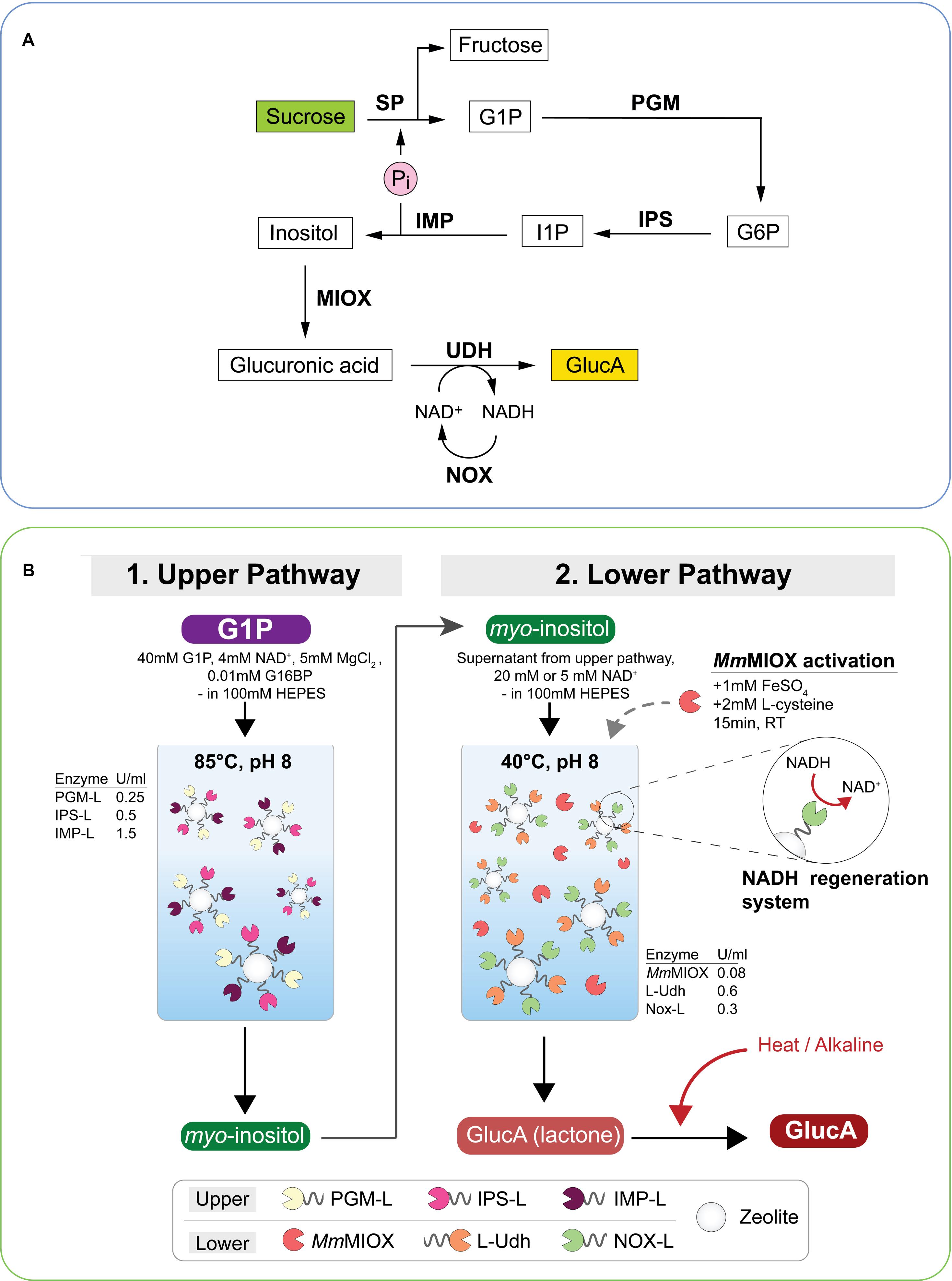
Figure 3. Schematic representation of glucaric acid (GlucA) synthesis using multi-enzyme pathways. (A) GlucA production from sucrose using mesophilic enzymes. SP, sucrose phosphorylase; PGM, phosphoglucomutase; IPS, inositol 1-phosphate synthase; IMP, myo-inositol monophosphatase; MIOX, myo-inositol oxygenase; UDH, uronate dehydrogenase; NOX, NADH oxidase; Pi, inorganic phosphate. Production of inositol by IMP produces Pi which is recycled in the first step of the reaction. Adapted from Su et al. (2019). (B) Production of GlucA from glucose-1-phosphate (G1P) using a two-batch reaction of the upper and lower pathway. Mainly thermophilic enzymes were selected for the pathway. Enzyme concentrations are shown in U mL– 1, MIOX was pre-activated with Fe and Cys; the immobilized NAD+ regeneration system (Nox-L) and conversion of glucaro-1,4-lactone to GlucA are shown. Enzymes carrying a silica-binding peptide (L) were immobilized onto zeolite while MmMIOX was used as free enzyme. PGM-L, phosphoglucomutase; IPS-L, myo-inositol-3-phosphate synthase; IMP-L, inositol-1-monophosphatase; LUdh, uronate dehydrogenase; Nox-L, NADH oxidase; MmMIOX, myo-inositol oxygenase; G16BP, glucose 1,6-bisphosphate; RT, room temperature. Reproduced with permission from Petroll et al. (2020) 2018 Elsevier Inc.
Petroll et al. (2020) reported the cell-free enzymatic production of glucaric acid from glucose-1-phosphate with a titre of 14.1 mM using free enzymes and 8.1 mM using immobilized enzymes with a total reaction time of 10 h in a two-pot reaction system incorporating a cofactor regeneration system (Figure 3B). The highest productivities, 0.30 g h–1 L–1 for free enzymes and 0.170 g h–1 L–1 for immobilized enzymes, so far were reported for glucaric acid production using a synthetic enzyme pathway. The two-pot reaction was necessary because of the comparative thermal instability of the Mus musculus MIOX, the Fulvimarina pelagi uronate dehydrogenase and the Lactobacillus rhamnosus NADPH oxidase enzymes as part of the synthetic pathway.
Thaore et al. (2020) have published a techno-economic and a life cycle analysis of the production of glucaric acid from corn stover by oxidation of glucose and showed at least in silico, that a commercially competitive product could be produced in each pathway. Azeotropic concentration at a low temperature of the glucaric acid was proposed for the enzymatic pathway to overcome the occurrence of D-glucaro-1,4-lactone but Petroll et al. (2020) demonstrated that it could be converted to glucaric acid by a simple heat treatment under alkaline conditions (Figure 3B).
A number of other systems utilizing synthetic biology, multi-enzyme biocatalysis and cell-free synthesis aimed at the production of several intermediates and precursors have been described. We review an incomplete range of these systems and their applications. Terpenes are a large class of industrially relevant substances that have found uses such as flavors, fragrances vitamins and pharmaceuticals. Many are plant-derived and suffer from low yields in nature. Both E. coli and Saccharomyces cerevisiae have been engineered genetically to produce a variety of mono-, di, and sesqui-terpenes but the yields necessary for commercial success have not been achieved, partially due to toxicity barriers (reviewed by Korman et al., 2017). A cell-free system was constructed consisting of 27 enzymes for the conversion of glucose to monoterpenes such as pinene, limonene and sabinene with the concurrent regeneration of NADPH and ATP. Conversion yields of more than 95% and titres of more than 15 g L–1 were achieved (Korman et al., 2017). The pathway was constructed from enzymes of the glycolytic pathway feeding acetyl-CoA into the 8 pathway as well as some auxiliary enzymes to regenerate cofactors. Different mono-terpenes were produced by using alternative terpene synthases in the final step of the combined pathways. The enzymes were cloned and expressed individually using His-tags and combined in suitable ratios for maximal synthesis as an example of multi-enzyme catalysis. The synthesis of limonene from glucose continued for at least 5 days until the glucose was exhausted.
A simplification in the production of limonene was reported by Dudley et al. (2019) who used a mixture of cell-free lysates from E. coli each containing a single over-expressed enzyme in the pathway using mevalonate as substrate and then using glucose plus native enzymes in the extracts to convert glucose to mevalonate plus the similar addition of lysates containing three additional enzymes. The crude lysates supported the complex biochemical metabolism involved and the general approach has promise for construction and characterisation of other pathways. Using a similar approach, Moore et al. (2017b) described the one-pot synthesis involving a purified five enzyme cascade from the substrate tyrosine to the valuable chemical raspberry ketone as the result of balancing the key cofactor, coenzyme A, to give an economically valuable yield unobtainable in whole cell catalysis with E. coli and S. cerevisiae.
Pinene production was reported to be improved by a combinatorial approach using tolerance, evolution and modular coculture engineering using a cell-free system that was constructed using the upstream module from E. coli strain MEVI and the downstream module from E. coli strain PINE (Niu et al., 2020). On scale-up it was found that the surface area to volume ratio altered cell-free synthesis of pinene. The titre obtained was lower than that of Korman et al. (2017) but the productivity was 18% higher. The fed batch mode could not be used to increase pinene production due to the formation of precipitate in the cell-free reaction system but immobilization of the crude lysate may enhance its production.
An early attempt at the simple in vitro production of the next generation fuel, n-butanol, was reported by Krutsakorn et al. (2013) who took advantage of the thermostability of enzymes from common thermophiles when expressed in the mesophilic host E. coli which allowed partial purification by exposure to high temperatures. They designed a heterologous, oxygen-insensitive pathway for the direct conversion of glucose to n-butanol by producing sixteen catalytic modules that were balanced for consumption and regeneration of ADP and ATP and NAD+ and NADH with flux optimisation for each enzyme. Most enzymes were cloned from Thermus thermophilus but others were from archaea such as Thermococcus and Pyrococcus with a weak point (in retrospect) being the use of a Sulfolobus dehydratase in the 3-hydroxybutyryl-CoA to crotonyl-CoA step. However, when optimized, the pathway produced n-butanol at a molar yield of 82%.
The replacement of fossil fuels in the production of high value chemicals was examined by Sutiono et al. (2020) with reference to cell-free bio-transformations of D-glucose and glycerol to pyruvate as a precursor for alcohols, amino acids and other building blocks. They concentrated on the slow dehydration step of D-glycerate to pyruvate catalyzed by Sulfolobus solfataricus previously incorporated into enzyme cascades and searched for alternative enzymes with higher activities, finding two distinct enzymes with greater activity and higher turnover numbers than the enzyme from S. solfataricus. Replacement of this enzyme by the dehydratase from Paralcaligenes ureilyticus led to a 10-fold increase in total production at 92% theoretical yield and was compatible with glycerol as a substrate eliminating a bottleneck with the enzyme cascade. This research demonstrated again the value of combining heterologous enzymes (plus several auxiliary enzymes) in a synthetic cascade to provide greater yields of the final product which were enabled by multi-enzyme catalysis using partially purified catalysts.
Other researchers have focused on assembling enzymatic catalytic cascades from natural and non-natural sources for the conversion of waste products into value-added biochemicals or precursors. For example, a cell-free reaction cascade was constructed to convert spent coffee grounds into lactic acid (Kopp et al., 2019a). Four thermostable enzymes were combined in a one-pot reaction which gave an initial yield of 4.4 mM lactic acid with scope for further improvement (Figure 1A and see section “Substrates for Biomass Transformation”). Biologically based bioplastics to replace those derived from petroleum have been a source of interest for many research groups. Kelwick et al. (2018) described an E. coli-based cell-free system for the production of polyhydroalkanoate (PHA) plastics from whey permeate in an enzyme cascade with constitutive phaCAB operon products, β-galactosidase and acetyl-CoA provided by glycolysis and aided by a plasmid carrying T7 RNA polymerase. The authors note that the biodegradable PHA plastics are more expensive to produce than the petroleum-sourced products and suggested that the system proposed would be valuable at the microplate level in the search for more efficient producer combinations (see section “Cell-Free Enzyme Pathways”).
The commodity chemical 2,3-butanediol (2,3-BD) has applications in the manufacture of several products including inks, perfumes, explosives, polymers, paint, flavorings, and pharmaceuticals. It is difficult to synthesize 2,3-BD chemically as a single chiral form so that biological pathways that produce only one isoform are preferred. Kay and Jewett (2015) developed a pathway for the production of 2,3-BD from pyruvate in three steps that required only NADH as cofactor. They created strains of E. coli with heterologous pathways for 2,3-BD from pyruvate using bacterial crude cell lysates containing all enzymes necessary for the conversion. Recent advances in purified cell-free systems enabled a continuous supply of reducing equivalents and ATP, producing high titres but they might not be sustainable economically (Korman et al., 2014,2017). The crude lysate of the engineered E. coli reached 82 g L–1 titre in a 30 h fed-batch mode with 11.3 g L–1 h–1 peak volumetric productivity (Kay and Jewett, 2015). They identified conditions tolerated by the crude lysate of E. coli but toxic to whole cells such as antibiotics, polar solvents and pre-treated biomass hydrolysates. Although polar solvents affected enzyme stability in the biosynthetic reaction, the rate of 2,3-BD production is better than in the equivalent E. coli strains. Lower hydrophobic compounds had better tolerance at a greater fraction of total volume than the live E. coli strains. Pre-treated biomass hydrolysate samples were used in place of glucose and showed a 25% decrease in response to lignocellulose-derived toxins and lignocellulose-derived toxins like p-coumaric and ferulic acid, 5-hydroxymethyl-2-furaldehyde (HMF), coumaroyl and feruloyl amides were tolerated better by cell-free systems rather than intact cells. The conversion of cellodextrins from biomass into high energy phosphorylated oligosaccharides such as glucose-1-phosphate and glucose-6-phosphate in stoichiometric amounts was reported by Meng et al. (2018) using a three-enzyme cascade of a cell-free phosphorolytic enzyme system. They demonstrated the production of high concentrations of inositol from cellodextrins and showed that the cell-free system was not affected by toxins that limited production in whole cells of E. coli.
Problems of Scale-Up in Cell-Free Synthesis
Cell-free synthesis systems and pathways have been used extensively in the research and developmental areas but once the isolation of an improved form of the enzyme or pathway has been generated, commercial amounts usually are produced in vivo by traditional methods (reviewed in The Economist, 2017a, b; Rolf et al., 2019; The Economist, 2019). Only rarely have the results been published of scale up experiments. For example, Fujiwara and Doi (2016) achieved production of green fluorescent protein at the 9 liter level at 0.5 mg mL–1 with the expectation that the procedure could be scaled up to hundreds of liters. Researchers at Sutro Pharma Inc (South San Francisco, CA, United States) reported the linear scalability of their procedure to 100 L at a yield of ∼0.7 mg mL–1 for the synthesis of human granulocyte-macrophage colony-stimulating factor (Zawada et al., 2011). This development provides the cost-effective opportunity for the production of human therapeutic proteins and further scale up may not be essential [reviewed by Ogonah et al. (2017)].
The biotransformation of starch to myo-inositol at an industrial scale was reported by You et al. (2017). Myo-inositol is widely used in the pharmaceutical, cosmetic and food and feed industries and is produced mainly by chemical hydrolysis of phytate with accompanying problems of waste disposal, substrate and product isolation costs. Accordingly, the more “green” approach used partially purified hyperthermophilic enzymes to construct a multi-enzyme module that could convert starch to myo-inositol in a one pot reaction (Figure 1B). The catalytic cascade was composed of four enzymes from archaea and was supplemented by two other hyperthermophilic enzymes and was shown to be scalable to a 20,000 L reactor (You et al., 2017). A similar approach but involving some different sources of enzymes from hyperthermophiles was employed to catalyze synthesis of myo-inositol from sucrose (Zhong et al., 2017b). They developed a thermal cycling cascade using the less temperature-resistant sucrose phosphorylase immobilized on cellulose carrying magnetic nanoparticles. The cascade consisted of four enzymes without auxiliary factors and achieved a high molar yield of 0.98 mol of product per mol of sucrose. This study appeared to be a laboratory-based proof of concept and there was no report of large scale production as in the previous account from this group. In a recent study, Cheng et al. (2019) produced myo-inositol from D-xylose, an abundant sugar in lignocellulose biomass. They constructed an NAD(P)-free cascade involving eleven enzymes from hyperthermophiles that converted the 5-carbon ring of the substrate to the 6-carbon ring of the product, with the promiscuity of Thermotoga maritima xylulokinase allowing replacement of ATP by polyphosphate for xylulose phosphorylation (Figure 1C). The conversion efficiency from xylose to inositol was 96.6% at 70°C and the authors suggested that the enzyme cascade could be incorporated into the co-production of a value-added biochemical by biorefineries involved in ethanol production.
Commercialisation
A number of start-up companies have been set up in the cell-free synthetic biology space. We have evaluated them as far as possible from information on their web sites, where available. In addition, there are a number of mature companies that are present but most are contributing in the Information Technology (IT) area, in library production and screening and the provision of components to the start-ups in their discovery and laboratory demonstration areas. For example, the collaboration between Labcyte2, Arzeda3 and TeselaGen4 and Twist Bioscience5 has provided integration of leading technologies in DNA synthesis, liquid handling, and software for protein design as components for an assembly line for workflow in start-up companies. Genomatica6 has described the production of 1,4-butanediol in whole cell catalysis with E. coli (Yim et al., 2011; Bugard et al., 2016; Culler, 2016) but has tested and endorsed the use of cell-free extracts (termed TX-TL reactions) as a discovery and screening program. However, they have a number of products such as 1,3-butylene glycol, polyamide intermediates, and long chain chemicals developed for partners that have not moved to commercial-scale use of cell-free protein expression. Another major player is Sutro Biopharma (already referred to), who have been producing antibodies for cancer treatment in the form of conjugates using their system XpressCF+ to incorporate a non-natural amino acid, resulting in a protein that can be conjugated to one or more chemicals of interest in a site-specific manner to give homogenous single-species protein-drug conjugates. XpressCF+ has been used to make Sutro’s clinical ADC products, STRO-001 and STRO-002, in which a cytotoxin is conjugated to an antibody containing non-natural amino acid by cell-free synthesis reported to be at the 1,000 liter scale7.
Tierra8 have developed multiple cell-free systems representing and including Gram-positive, Gram-negative, and eukaryotic species. They have combined their multiple cell-free systems with automation, metagenomics, and reiterative learning. Tierra claim that they can scale production of particular proteins from the micro-scale (μL or μg) to testing scale (mL or mg). The most promising commercial proteins are claimed to be producible at the (L or kg) scale but no details were provided and they appear to be largely in the discovery phase.
Greenlight Biosciences9 have focused on RNA interference (RNAi) where an RNA molecule stops genes from being expressed, interfering with their message, as applied to agricultural pest control. The RNA-based solution is applied to a plant to silence specific genes in the pest which is killed while the plant stays healthy and there is no negative impact on the environment. Greenlight’s sprayable, double-stranded RNA solution can be used in agriculture to control pests (insects, weeds and fungi) in a highly specific manner. It has been produced by depolymerising RNAi by a nuclease, the products phosphorylated by thermophilic nucleotide kinases and then polymerised into double-stranded RNA using a DNA template coding for the desired protein. The procedure can be programmed to produce single or double-stranded RNA that can be used to combat plant pathogens (Blake et al., 2017). One product has been tested in field trials where the Colorado Potato Beetle is active, and efficiency has been confirmed in laboratory and greenhouse insect-feeding assays. While their first RNA product targets an insect larva, the flexibility of RNA technology has allowed the building of a pipeline of products other pest and disease problems facing farmers. All of the necessary thermophilic enzymes can be produced in a single E. coli recombinant and all reactions occur in a cell-free melange although they are not formerly organized into a cascade. In addition, GreenLight has been reported as utilizing its Cell-Free Bioprocessing platform to develop other RNA-based products. For example, GreenLight, is using its own E. coli cell-free system to produce industrial quantities of an undigestible analog of ribose for use in zero-calorie beverages and claims it can make thousands of liters of solution of this sugar at a time.
Several start-ups are pursuing CRISPR-Cas-based diagnostic tests (Sherlock Biosciences10; Mammoth Biosciences11) although the reactions to prepare the components only fit the description of being multi-enzyme reactions (Chiu, 2018). Arbor Biosciences12 is another new company whose aim appears to be the provision of tools for the discovery phase to other companies or partners and its capabilities are largely directed toward laboratory-based outcomes. Aralez Bioscience12 uses a cell-free system but with only a single class of enzyme that can perform condensation of a substituted indole with a β-hydroxy amino acid, such as serine or threonine, to produce an enantiopure tryptophan analog with water as the only by-product. In addition to indoles, the enzymes accept nitrogen and sulfur nucleophiles to make other classes of non-canonical amino acids. Cell-free synthesis has extended to membrane proteins that represent up to 60% of all pharmaceutical targets and Synthelis’ proprietary cell-free technology13 has been specifically designed to overcome production limitations, providing a large amount of functional membrane protein imitating a living cell environment.
Most research activity has been centered on E. coli but other systems have been developed including S. cerevisiae, various Streptomyces species, Bacillus subtilis, Gluconacetobacter hansenii, CHO cells, and HeLa cells to promote sulfide-bond formation, protein folding and other post-translational modifications (Hodgman and Jewett, 2013; Brodel et al., 2014; Kelwick et al., 2016; Li et al., 2017; Zhou et al., 2017). The broad picture for CFS in the production of biotherapeutics has been reviewed by Ogonah et al. (2017) who conclude that CFS may provide a nimbler system suited to distributed manufacture of single dose therapeutic proteins than traditional cell-based platforms.
Whether cell-free biotechnology can fully replace the current fermentation process that relies on genetically modified organisms for producing chemicals is arguable. Fermentation is an accepted and established industrial technique and has been used by humans for at least 12,000 years. However, current advances in cell-free biotechnology implies that for some applications the utility of the biological cell may be exhausted.
Conclusion and Perspectives
Cell-free protein expression has been revitalized over the past several decades by a number of technical advances since the early demonstration that whole cell integrity is not required for transcription and translation to occur (comprehensively reviewed in Katzen et al., 2005; Lu, 2017; Bowie et al., 2020; Silverman et al., 2020 and a number of other contributions). The current literature is also replete with top-down and bottom up suggestions on the way forward for CFS in the synthesis of bio-commodities (for example, Lu, 2017; Arbige et al., 2019; Bowie et al., 2020; Libicher et al., 2020). There are many valuable reviews that summarize the status of multi-enzyme arrays and cascades and their possible immobilization all mention the perceived advantages of CFS (for example, Katzen et al., 2005; Smith et al., 2014; Guo et al., 2017; Sperl and Sieber, 2018; Chemla et al., 2019; Hwang and Lee, 2019; Tinafar et al., 2019; Bowie et al., 2020; Giannakopoulou et al., 2020; Silverman et al., 2020). In all cases, considerable faith has been put in the prospect of present and future genetic manipulations to improve the performance of various proteins.
Most of the successful proof of concept reports have used enzymes sourced from thermophiles by discovery techniques and it is implied that they could be improved by judicious genetic engineering. This proposition may be true for directed evolution at the cost of substantial amounts of time and effort but most research is based on acceptance of the Classical Model of enzyme activity and inactivation as taught from undergraduate years. Recent results and the proposal of a new model aiming to explain the effect of temperature on enzyme activity may improve our understanding of enzymes in many areas including enzyme prospecting, enzyme engineering and enzyme production in a bioreactor. Little emphasis has been placed on the Equilibrium Model since its postulation (Daniel et al., 2001, 2010; Daniel and Danson, 2013) which predicts quite a different behavior of enzymes compared to the Classical Model, particularly in biotechnological applications where yield is a major factor (Eisenthal et al., 2006). It is highly desirable for many applications that the enzyme should be thermostable, particularly if recovery and repeated use are envisaged. The widely used Classical Model assumes that an increase in temperature results in increased enzyme activity until a temperature is reached or exceeded where activity is lost due to enzyme (protein) denaturation. This model is challenged by the Equilibrium Model which accounts for loss of enzymatic activity as temperature increases and before complete thermally induced protein denaturation (Peterson et al., 2004; Eisenthal et al., 2006; Daniel and Danson, 2010, 2013; Daniel et al., 2010). The model is based on the notion that the more flexible active site of an enzyme will be affected by increased temperatures before the whole protein is denatured. Accordingly, the model assumes an intermediate enzyme species which is catalytically inactive but non-denatured. Based on the existence of this inactive intermediate, the Equilibrium Model introduces the new thermal parameter Teq, which represents the temperature at which the concentration of the active and inactive forms of the enzyme, are equal (Peterson et al., 2004). In the Classical Model, the determination of the temperature optimum (Topt) of an enzyme relies directly on the length of the assay because thermal denaturation is time dependent (Eisenthal et al., 2006). As such, any change in the length of the assay will directly affect the apparent temperature optimum of the enzyme. Based on the Equilibrium Model, both the thermal stability and activity (via Teq) need to be considered for screening of new thermostable enzymes rather than by selecting only for thermal activity. The Classical Model rightly continues to be used to analyze the performance of enzymes and may assist in the improvement of enzymes as appropriate but the results of the Daniel-Danson group have made it clear that increased thermostability cannot be viewed in isolation where improved performance is required for an enzyme and the Equilibrium Model provides a second way in which temperature affects enzyme activity, probably at the active site. Although stable enzymes are important in CFS, it should be noted that so far, the results for their stability improvement have been disappointing in general (as they have not included Teq considerations, as discussed in Daniel et al., 2010; Lee et al., 2013) and the observation of increased stability but decreased activity has been a frequent (and usually unpublished) outcome although there are some successes (e.g., Shakeel et al., 2019).
The particular advantage of CFS in overcoming many of the constraints experienced with whole cell enzyme production has been emphasized by most commentators. However, when one examines the frequently used example of biomass (of several varieties) as a substrate for biofuels and commodity chemicals there is little evidence of scale up. In the experiments of the production of myo-inositol, there was the report of production at a 20,000 L level (You et al., 2017), but apart from the results reported by Sutro Inc on the production of biotherapeutics at the hundreds of liters scale, most research findings are at the laboratory bench level. If biomass is to be the substrate for biofuels, enzyme requirements are in the thousands of liters class to be economically competitive with fossil fuel-derived products. Accordingly, a concerted research effort is required to increase scale up and to shift from batch to continuous synthesis.
Other areas of significance in lowering the cost of protein production include establishment of a highly efficient cofactor system for energy regeneration and wider examination of the use of RNA-derived solutions for areas other than pest control. Other promising developments are the formation of a large self-replicating synthetic genomes as plasmids to enable the synthesis of multiple proteins as a self-replenishing translational-transcriptional system (Libicher et al., 2020) and improved immobilization of enzymes in cascades with low-cost matrices and without the necessity of chemical reactions that lead to (partial) inactivation. Earlier research encompassing microporous agarose beads and enzyme immobilization by either metal coordination or aldehyde chemistry are reviewed by López-Gallego (2019) but other simpler and effective approaches have been suggested (Care et al., 2015). There seems to be a considerable time gap between demonstration on the laboratory bench and commercial acceptance, demonstration and production, at least in part because of the material cost of scale up. It would appear that successful CFS prototyping experiments finally end up back in whole cell fermentations which are more familiar to commercial biotechnology.
Another emerging future trend is the development of enzymes with genetically encoded unnatural amino acids (reviewed by Burgener et al., 2020; Drienovská and Roelfes, 2020 and also see Yu et al., 2018) and put into practice by Zhou and Roelfes (2020) who inserted a non-natural amino acid at the catalytic site of the transcription factor LmrR and with a complex containing copper, was able to activate reaction partners in the Michael addition reaction which creates carbon-carbon bonds. These experiments are single enzyme reactions at present but there is discussion on their incorporation into enzyme cascades which will probably include CFS.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
SS is supported by an International Macquarie University Research Excellence Scholarship (iMQRES).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^ www.sutrobio.com
- ^ www.labcyte.com
- ^ www.arzeda.com
- ^ www.teselagen.com
- ^ www.twistbioscience.com
- ^ www.genomatica.com
- ^ www.economist.com/leaders/2017/05/04/the-remarkable-promise-of-cell-free-biology
- ^ www.tierrabiosciences.com
- ^ www.greenlightbiosciences.com
- ^ www.sherlock.bio
- ^ www.mammoth.bio
- ^ arborbiosciences.com
- ^ www.synthelis.com
References
Aden, A., Ruth, M., Ibsen, K., Jechura, J., Neeves, K., Sheehan, J., et al. (2002). Lignocellulosic Biomass to Ethanol Process Design and Economics Utilizing Co-Current Dilute Acid Prehydrolysis and Enzymatic Hydrolysis for Corn Stover. Golden, CO: National Renewable Energy Lab.
Alissandratos, A., Hartley, C. J., French, N. G., Kim, H.-K., Allen, S., Estavillo, G. M., et al. (2019). One-pot multienzymatic transformation of NH3, CO2, and ornithine into the organic nitrogen plant fertilizer citrulline using a single recombinant lysate of E. coli. ACS Sustain. Chem. Eng. 7, 8522–8529. doi: 10.1021/acssuschemeng.9b00301
Andexer, J. N., and Richter, M. (2015). Emerging enzymes for ATP regeneration in biocatalytic processes. ChemBioChem 16, 380–386. doi: 10.1002/cbic.201402550
Anuar, M. R., and Zuhairi, A. (2016). Challenges in biodiesel industry with regards to feedstock, environmental, social and sustainability issues: a critical review. Renew. Sustain. Energy Rev. 58, 208–223. doi: 10.1016/j.rser.2015.12.296
Arbige, M. V., Shetty, J. K., and Chotani, G. K. (2019). Industrial enzymology: the next chapter. Trends Biotechnol. 37, 1355–1366. doi: 10.1016/j.tibtech.2019.09.010
Asano, N., Oseki, K., Tomioka, E., Kizu, H., and Matsui, K. (1994). N-containing sugars from Morus alba and their glycosidase inhibitory activities. Carbohydr. Res. 259, 243–255. doi: 10.1016/0008-6215(94)84060-1
Ayoubi-Joshaghani, M. H., Dianat-Moghadam, H., Seidi, K., Jahaban-Esfahalan, A., Zare, P., and Jahanban-Esfhalan, R. (2020). Cell-free protein synthesis: the transition from batch reactions to minimal cells and microfluidic devices. Biotechnol. Bioeng. 117, 1204–1229. doi: 10.1089/ind.2019.29187.nla
Aznar, N. L. (2019). Sustainable fibers for textile applications. Ind. Biotechnol. 15, 290–292. doi: 10.1089/ind.2019.29187.nla
Babich, L., Hartog, A. F., van Hemert, L. J. C., Rutjes, F. P. J. T., and Wever, R. (2012). Synthesis of carbohydrates in a continuous flow reactor by immobilized phosphatase and aldolase. ChemSusChem 5, 2348–2353. doi: 10.1002/cssc.201200468
Babich, L., van Hemert, L. J. C., Bury, A., Hartog, A. F., Falcicchio, P., van der Oost, J., et al. (2011). Synthesis of non-natural carbohydrates from glycerol and aldehydes in a one-pot four-enzyme cascade reaction. Green Chem. 13, 2895–2900. doi: 10.1039/C1GC15429J
Beauchamp, J., and Vieille, C. (2015). Activity of select dehydrogenases with sepharose-immobilized N6-carboxymethyl-NAD. Bioengineered. 6, 106–110. doi: 10.1080/21655979.2014.1004020
Beer, B., Pick, A., and Sieber, V. (2017). In vitro metabolic engineering for the production of α-ketoglutarate. Metab. Eng. 40, 5–13. doi: 10.1016/j.ymben.2017.02.011
Benítez-Mateos, A. I., Contente, M. L., Velasco-Lozano, S., Paradisi, F., and López-Gallego, F. (2018). Self-sufficient flow-biocatalysis by coimmobilization of pyridoxal 5′-phosphate and ω-transaminases onto porous carriers. ACS Sustainable Chem. Eng. 6, 13151–13159. doi: 10.1021/acssuschemeng.8b02672
Benítez-Mateos, A. I., San Sebastian, E., Ríos-Lombardía, N., Morís, F., González-Sabín, J., and López-Gallego, F. (2017). Asymmetric reduction of prochiral ketones by using self-sufficient heterogeneous biocatalysts based on NADPH-dependent ketoreductases. Chemistry 23, 16843–16852. doi: 10.1002/chem.201703475
Bilal, M., and Iqbal, H. M. N. (2019). Sustainable bioconversion of food waste into high-value products by immobilized enzymes to meet bio-economy challenges and opportunities – A review. Food Res. Int. 123, 226–240. doi: 10.1016/j.foodres.2019.04.066
Blake, W. J., Cunningham, D. S., MacEachran, D., Gupta, M., and Abshire, J. (2017). Cell Free Production of Nucleic Acid. U.S. Patent 20170292138A1. Geneva: World Intellectual Property Organization.
Blank, L. M., Narancic, T., Mampel, J., Tiso, T., and O’Connor, K. (2020). Biotechnological upcycling of plastic waste and other non-conventional feedstocks in a circular economy. Curr. Opin. Biotech. 62, 212–219. doi: 10.1016/j.copbio.2019.11.011
Bornscheuer, U. T. (2016). Biocatalysis: successfully crossing boundaries. Angew. Chim. Int. Edn. 55, 4372–4373. doi: 10.1002/anie.201510042
Bornscheuer, U. T. (2018). The fourth wave of biocatalysis is approaching. Philos. Trans. A Math. Phys. Eng. Sci. 376:20170063. doi: 10.1098/rsta.2017.0063
Bornscheuer, U. T., Huisman, G. W., Kazlauskas, R. J., Lutz, S., Moore, J. C., and Robins, K. (2012). Engineering the third wave of biocatalysis. Nature 485, 185–194. doi: 10.1038/nature11117
Bowie, J. U., Sherkhanov, S., Korman, T. P., Valliere, M. A., Opgenorth, P. H., and Liu, H. (2020). Synthetic biochemistry: the bio-inspired cell-free approach to commodity chemical production. Trends Biotechnol. 38, 766–778. doi: 10.1016/j.tibtech.2019.12.024
Britton, J., Majumdar, S., and Weiss, G. A. (2018). Continuous flow biocatalysis. Chem. Soc. Rev. 47, 5891–5918. doi: 10.1039/C7CS00906B
Brodel, A. K., Sonnabend, A., and Kubick, S. (2014). Cell-free protein expression based on extracts from CHO cells. Biotechnol. Bioeng. 111, 25–36. doi: 10.1002/bit.25013
Bugard, A., Burk, M. J., Osterhout, R., Van Dien, S., and Yim, H. (2016). Development of a commercial scale process for production of 1,4-butanediol from sugar. Curr. Opin. Biotechnol. 42, 118–125. doi: 10.1016/j.copbio2016.04.016
Burgener, S., Luo, S., McLean, R., Miller, T. E., and Erb, T. J. (2020). A roadmap towards integrated catalytic systems of the future. Nat. Catal. 3, 186–192. doi: 10.1038/s41929-020-0429-x
Caetano, N. S., Silva, V. F. M., and Mata, T. M. (2012). Valorization of coffee grounds for biodiesel production. Chem. Eng. Trans. 26, 267–272. doi: 10.3303/CET1226045
Cai, Q., Hanson, J. A., Steiner, A. R., Tran, C., Masikat, M. R., Chen, R., et al. (2015). A simplified and robust protocol for immunoglobulin expression in Escherichia coli cell-free protein synthesis systems. Biotechnol. Prog. 31, 823–831. doi: 10.1002/btpr.23103
Campos-Vega, R., Loarca-Piña, G., Vergara-Castañeda, H. A., and Oomah, B. D. (2015). Spent coffee grounds: a review on current research and future prospects. Trends Food Sci. Technol. 45, 24–36. doi: 10.1016/j.tifs.2015.04.012
Care, A., Bergquist, P. L., and Sunna, A. (2015). Solid-binding peptides: smart tools for nanobiotechnology. Trends Biotechnol. 33, 259–268. doi: 10.1016/j.tibtech.2015.02.005
Carlson, E. D., Gan, R., Hodgman, C. E., and Jewett, M. C. (2012). Cell-free protein synthesis: applications come of age. Biotechnol. Adv. 30, 1185–1194. doi: 10.1016/j.biotechadv.2011.09.016
Carus, M., and Dammer, L. (2018). The circular bioeconomy-concepts, opportunities and limitations. Ind. Biotech. 14, 83–91. doi: 10.1089/ind.2018.29121.mca
Castillo, J. A., Calveras, J., Casas, J., Mitjans, M., Vinardell, M. P., Parella, T., et al. (2006). Fructose-6-phosphate aldolase in organic synthesis: preparation of D-fagomine, N-alkylated derivatives, and preliminary biological assays. Org. Let. 8, 6067–6070. doi: 10.1021/ol0625482
Chemla, Y., Ozer, E., Shaferman, M., Zaad, B., Dandela, R., and Alfonta, L. (2019). Simplified methodology for a modular and genetically expanded protein synthesis in cell-free systems. Synth. Syst. Biotechnol. 4, 189–196. doi: 10.1016/j.synbio.2019.10.002
Cheng, K., Zheng, W., Chen, H., and Job Zhang, Y.-H. P. (2019). Upgrade of wood sugar D-xylose to a value-added nutraceutical by in vitro metabolic engineering. Metab. Eng. 52, 1–8. doi: 10.1016/j.ymben.2018.10.007
Cherubini, F. (2010). The biorefinery concept: using biomass instead of oil for producing energy and chemicals. Energ. Convers. Manage. 51, 1412–1421. doi: 10.1016/j.enconman.2010.01.015
Chiu, C. (2018). Cutting-edge infectious disease diagnostics with CRISPR. Cell Host Microbe. 23, 702–704. doi: 10.1016/j.chom.2018.05.016
Chong, S. (2014). Overview of cell-free protein synthesis: historic landmarks, commercial systems, and expanding applications. Curr. Protoc. Mol. Biol. 108, 16.30.1–16.30.11. doi: 10.1002/0471142727.mb1630s108
Clomburg, J. M., Crumbley, A. M., and Gonzalez, R. (2017). Industrial biomanufacturing: the future of chemical production. Science 355, 38–47.
Culler, S. (2016). A bioengineering platform to industrialize biotechnology. Chem. Eng. Prog. 112, 42–51.
Daniel, R. M., and Danson, M. J. (2010). A new understanding of how temperature affects the catalytic activity of enzymes. Trends Biochem. Sci. 35, 584–591. doi: 10.1016/j.tibs.2010.05.001
Daniel, R. M., and Danson, M. J. (2013). Temperature and the catalytic activity of enzymes: a fresh understanding. FEBS Lett. 587, 2738–2743. doi: 10.1016/j.febslet.2013.06.027
Daniel, R. M., Danson, M. J., and Eisenthal, R. (2001). The temperature optima of enzymes: a new perspective on an old phenomenon. Trends Biochem. Sci. 26, 223–225. doi: 10.1016/S0968-0004(01)01803-5
Daniel, R. M., Peterson, M. E., Danson, M. J., Price, N. C., Kelly, S. M., Monk, C. R., et al. (2010). The molecular basis of the effect of temperature on enzyme activity. Biochem. J. 425, 353–360. doi: 10.1042/BJ20091254
DeBoer, J., Panwar, R., Kozak, R., and Cashore, B. (2020). Squaring the circle: refining the competitiveness logic for the circular bioeconomy. Forest Pol. Econom. 110, 101858. doi: 10.1016/j.forpol.2019.01.003
DeSantis, G., Wong, K., Farwell, B., Chatman, K., Zhu, Z., Tomlinson, G., et al. (2003). Creation of a productive, highly enantioselective nitrilase through gene site saturation mutagenesis (GSSM). J. Am. Chem. Soc. 125, 11476–11477.
Devries, J. K., and Zubay, G. (1967). DNA-directed peptide synthesis. II. The synthesis of the alpha-fragment of the enzyme beta-galactosidase. Proc. Natl. Acad. Sci. U.S.A 57, 1010–1012. doi: 10.1073/pnas.57.4.1010
Drienovská, I., and Roelfes, G. (2020). Expanding the enzyme universe with genetically encoded unnatural amino acids. Nat. Catal. 3, 193–202. doi: 10.1038/s41929-019-0410-8
Dudley, Q. M., Anderson, K. C., and Jewett, M. C. (2016). Cell-free mixing of Escherichia coli crude extracts to prototype and rationally engineer high-titer mevalonate synthesis. ACS Synth. Biol. 5, 1578–1588. doi: 10.1021/acssynbio.6b00154
Dudley, Q. M., Karim, A. S., Nash, C. J., and Jewett, M. C. (2020). Cell-free prototyping of limonene biosynthesis using cell-free protein synthesis. Metab. Eng. 61, 251–260. doi: 10.1016/j.ymben.2020.05.006
Dudley, Q. M., Nash, C. J., and Jewett, M. C. (2019). Cell-free biosynthesis of limonene using enzyme-enriched Escherichia coli lysates. Synth. Biol. 4:ysz003. doi: 10.1093/synbio/ysz003
Eisenthal, R., Peterson, M. E., Daniel, R. M., and Danson, M. J. (2006). The thermal behaviour of enzyme activity: implications for biotechnology. Trends Biotechnol. 24, 289–292. doi: 10.1016/j.tibtech.2006.05.004
El-Zahab, B., Donnelly, D., and Wang, P. (2008). Particle-tethered NADH for production of methanol from CO2 catalyzed by coimmobilized enzymes. Biotechnol. Bioeng. 99, 508–514. doi: 10.1002/bit.21584
Erickson, B. (2018). A rising tide of cellulosic ethanol production. Ind. Biotech. 14, 77–78. doi: 10.1089/ind.2018.29124.ber
FAO (2011). Global Food Losses and Food Waste: Extent, Causes and Prevention. Rome: Food and Agriculture Organization of the United Nations.
Fernandes, A. S., Mello, F. V. C., Thode Filho, S., Carpes, R. M., Honório, J. G., Marques, M. R. C., et al. (2017). Impacts of discarded coffee waste on human and environmental health. Ecotoxicol. Environ. Saf. 41, 30–36. doi: 10.1016/j.ecoenv.2017.03.011
Findrik, Z., and Vasić-Rački, D. (2007). Biotransformation of D−methionine into L−methionine in the cascade of four enzymes. Biotechnol. Bioeng. 98, 956–967. doi: 10.1002/bit.21501
France, S. P., Hepworth, L. J., Turner, N. J., and Flitsch, S. L. (2017). Constructing biocatalytic cascades: in vitro and in vivo approaches to de novo multi-enzyme pathways. ACS Catal. 7, 710–724. doi: 10.1021/acscatal.6b02979
Friehs, K. (2004). “Plasmid copy number and plasmid stability,” in New Trends and Developments in Biochemical Engineering, ed. T. Scheper (Berlin: Springer), 47–82. doi: 10.1007/b12440
Fu, J., Yang, Y. R., Johnson-Buck, A., Liu, M., Liu, Y., Walter, N. G., et al. (2014). Multi-enzyme complexes on DNA scaffolds capable of substrate channelling with an artificial swinging arm. Nat. Nanotechnol. 9, 531–536. doi: 10.1038/nnano.2014.100
Fujisawa, T., Fujinaga, S., and Atomi, H. (2017). An in vitro enzyme system for the production of myo-inositol from starch. Appl. Environ. Microbiol. 83:e00550-17. doi: 10.1128/AEM.00550-17
Fujiwara, K., and Doi, N. (2016). Biochemical preparation of cell extract for cell-free protein synthesis without physical disruption. PLoS One 11:e0154614. doi: 10.1371/journal.pone0154614
Gan, R., and Jewett, M. C. (2014). A combined cell-free transcription-translation system from for rapid and robust protein synthesis. Biotechnol. J. 9, 641–651. doi: 10.1002/biot.201300545
Gao, C., Li, Z., Zhang, L., Wang, C., Li, K., Ma, C., et al. (2015). An artificial enzymatic reaction cascade for a cell-free bio-system based on glycerol. Green Chem. 17, 804–807. doi: 10.1039/C4GC01685H
Gao, H., Tiwari, M. K., Kang, Y. C., and Lee, J.-K. (2012). Characterization of H2O-forming NADH oxidase from Streptococcus pyogenes and its application in l-rare sugar production. Bioorg. Med. Chem. Lett. 22, 1931–1935. doi: 10.1016/j.bmcl.2012.01.049
Gao, Q., Wang, X., Hu, S., Xu, N., Jiang, M., Ma, C., et al. (2019). High-yield production of D-1,2,4-butanetriol from lignocellulose-derived xylose by using a synthetic enzyme cascade in a cell-free system. J. Biotechnol. 292, 76–83. doi: 10.1016/j.jbiotec.2019.01.004
Garcia, D. C., Mohr, B. P., Dovgan, J. T., Hurst, G. B., Standaert, R. F., and Doktycz, M. J. (2018). Elucidating the potential of crude cell extracts for producing pyruvate from glucose. Synth. Biol. 3:ysy006. doi: 10.1093/synbio/ysy006
Geueke, B., Riebel, B., and Hummel, W. (2003). NADH oxidase from Lactobacillus brevis: a new catalyst for the regeneration of NAD. Enzym. Microb. Technol. 32, 205–211. doi: 10.1016/S0141-0229(02)00290-9
Giannakopoulou, A., Gkantzou, E., Polydera, A., and Stamaytis, A. (2020). Multienzymatic nanoassemblies: recent progress and applications. Trends Biotechnol. 38, 202–216. doi: 10.1016/j.tibtech.2019.07.010
Goldsmith, M., and Tawfik, D. S. (2017). Enzyme engineering: reaching the maximal catalytic efficiency peak. Curr. Opin. Struct. Biol. 47, 140–150. doi: 10.10016/j.sbi.2017.09.002
Gooday, G. W. (1990). “The ecology of chitin degradation,” in Advances in Microbial Ecology, Vol. 11, ed. K. C. Marshall (Boston: Springer), 387–430. doi: 10.1007/978-1-4684-7612-5_10
Grubbe, W. S., Rasor, B. J., Krüger, A., Jewett, M. C., and Karim, A. S. (2020). Cell-free styrene biosynthesis at high titers. Metab. Eng. 61, 89–95. doi: 10.1016/j.ymben.2020.05.009
Gruber, P., Marques, M. P. C., O’Sullivan, B., Baganz, F., Wohlgemuth, R., and Szita, N. (2017). Conscious coupling: the challenges and opportunities of cascading enzymatic microreactors. Biotechnol. J. 12:art1700030. doi: 10.1002/biot.201700030
Gumulya, Y., and Gillam, E. M. (2017). Exploring the past and the future of protein evolution with ancestral sequence reconstruction: the ‘retro’ approach to protein engineering. Biochem. J. 474, 1–19. doi: 10.1042/BCJ2016050
Guo, W., Sheng, J., and Feng, X. (2017). Minireview: in vitro metabolic engineering for bio-manufacturing of high-value products. Comput. Struct. Biotechnol. J. 15, 161–167. doi: 10.1016/j.csbj.2017.01.006
Gupta, A., Hicks, M. A., Manchester, S. P., and Prather, K. L. (2016). Porting the synthetic D-glucaric acid pathway from Escherichia coli to Saccharomyces cerevisae. Biotechnol. J. 11, 1201–1208. doi: 10.1002/biot.201500563
Guterl, J. K., Garbe, D., Carsten, J., Steffler, F., Sommer, B., Reiße, S., et al. (2012). Cell-free metabolic engineering: production of chemicals by minimized reaction cascades. ChemSusChem 5, 2165–2172. doi: 10.1002/cssc.201200365
Hanatani, Y., Imura, M., Taniguchi, H., Okano, K., Toya, Y., Iwakiri, R., et al. (2019). In vitro production of cysteine from glucose. Appl. Microbiol. Biotechnol. 103, 8009–8019. doi: 10.1007/s00253-019-10061-4
Hartley, C. J., French, N. G., Scoble, J. A., Williams, C. C., Churches, Q. I., Frazer, A. R., et al. (2017). Sugar analog synthesis by in vitro biocatalytic cascade: a comparison of alternative enzyme complements for dihydroxyacetone phosphate production as a precursor to rare chiral sugar synthesis. PLoS One 12:e0184183. doi: 10.1371/journal.pone.0184183
Hartley, C. J., Williams, C. C., Scoble, J. A., Churches, Q. I., North, A., French, N. G., et al. (2019). Engineered enzymes that retain and regenerate their cofactors enable continuous-flow biocatalysis. Nat. Catal. 2, 1006–1015. doi: 10.1038/s41929-019-0353-0
Hegde, S., Lodge, J. S., and Trabold, T. A. (2018). Characteristics of food processing wastes and their use in sustainable alcohol production. Renew. Sust. Energ. Rev. 81, 510–523. doi: 10.1016/j.rser2017.07.012
Hodgman, C. E., and Jewett, M. C. (2013). Optimized extract preparation methods and reaction conditions for improved yeast cell-free protein synthesis. Biotechnol. Bioeng. 110, 2643–2654. doi: 10.1002/bit.24942
Honda, K., Hara, N., Cheng, M., Nakamura, A., Mandai, K., Okano, K., et al. (2016). In vitro metabolic engineering for the salvage synthesis of NAD+. Metab. Eng. 35, 114–120. doi: 10.1016/j.ymben.2016.02.005
Honda, K., Kimura, K., Ninh, P. H., Taniguchi, H., Okano, K., and Ohtake, H. (2017). In vitro bioconversion of chitin to pyruvate with thermophilic enzymes. J. Biosci. Bioeng. 124, 296–301. doi: 10.1016/j.jbiosc.2017.04.013
Horton, B. (2019). Beyond food: animals and the bioeconomy. Ind. Biotechnol. 15, 282–283. doi: 10.1089/ind.2019.29186.bho
Huang, C. J., Lin, H., and Yang, X. (2012). Industrial production of recombinant therapeutics in Escherichia coli and its recent advancements. J. Ind. Microbiol. Biotechnol. 39, 383–399. doi: 10.1007/s10295-11-1082-9
Huang, R., Chen, H., Zhong, C., Kim, J. E., and Zhang, Y.-H. P. (2016). High-throughput screening of coenzyme preference change of thermophilic 6-phosphogluconate dehydrogenase from NADP+ to NAD+. Sci. Rep. 6:32644. doi: 10.1038/srep32644
Hull, S. R., Yang, B. Y., Venzke, D., Kulhavy, K., and Montgomery, R. (1996). Composition of corn steep water during steeping. J. Agric. Food Chem. 44, 1857–1863. doi: 10.1021/jf950353v
Hwang, E. T., and Lee, S. (2019). Multienzymatic cascade reactions via enzyme complex by immobilization. ACS Catal. 9, 4402–4425. doi: 10.1021/acscatal.8b04921
Imbert, E. (2017). Food waste valorization options: opportunities from the bioeconomy. Open Agric. 2, 195–204. doi: 10.1515/opag-2017-0020
International Coffee Organization (2019). Statistics Crop Year 2017. Available online at: http://www.ico.org/new_historical.asp?section=Statistics. (accessed 8 April 2020)
Jewett, M. C., Calhoun, K. A., Voloshin, A., Wuu, J. J., and Swartz, J. R. (2008). An integrated cell-free metabolic platform for protein production and synthetic biology. Mol. Syst. Biol. 4:220. doi: 10.1038/msb2008.57
Ji, X., Su, Z., Wang, P., Ma, G., and Zhang, S. (2015). Tethering of nicotinamide adenine dinucleotide inside hollow nanofibers for high-yield synthesis of methanol from carbon dioxide catalyzed by coencapsulated multienzymes. ACS Nano. 9, 4600–4610. doi: 10.1021/acsnano.5b01278
Johnson, D. T., and Taconi, K. A. (2007). The glycerin glut: options for the value-added conversion of crude glycerol resulting from biodiesel production. Environ. Prog. 26, 338–348. doi: 10.1002/ep.10225
Jung, D. H., Jung, J. H., Seo, D. H., Ha, S. J., Kweon, D. K., and Park, C. S. (2013). One-pot bioconversion of sucrose to trehalose using enzymatic sequential reactions in combined cross-linked enzyme aggregates. Bioresour. Technol. 130, 801–804. doi: 10.1016/j.biortech.2012.12.162
Karim, A. S., and Jewett, M. C. (2016). A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab. Eng. 36, 116–126. doi: 10.1016/j.ymben.2016.03.002
Karim, A. S., and Jewett, M. C. (2018). Cell-free synthetic biology for pathway prototyping. Methods Enzymol. 608, 31–57. doi: 10.1016/bs.mie.2018.04.029
Karim, A. S., Rasor, B. J., and Jewett, M. C. (2020). Enhancing control of cell-free metabolism through pH modulation. Synth. Biol. 5:ysz027. doi: 10.1093/synbio/ysz027
Katzen, F., Chang, G., and Kudlicki, W. (2005). The past, present and future of cell-free protein synthesis. Trends Biotechnol. 23, 150–156. doi: 10.1016/j.tibtech.2005.01.003
Kay, J. E., and Jewett, M. C. (2015). Lysate of engineered Escherichia coli supports high-level conversion of glucose to 2,3-butanediol. Metab. Eng. 32, 133–142. doi: 10.1016/j.ymben.2015.09.015
Keasling, J. D., Mendoza, A., and Baran, P. A. (2012). A constructive debate. Nature 492, 188–189. doi: 10.10338/492188a
Kelwick, R., Ricci, L., Chee, S. M., Bell, D., Webb, A. J., and Freemont, P. S. (2018). Cell-free strategies for enhancing the sustainable production of polyhydroxyalkanoates bioplastics. Syn. Biol. 3:ysy016. doi: 10.1093/synbio/7sy016
Kelwick, R., Webb, A. J., MacDonald, J. T., and Freemont, P. S. (2016). Development of a Bacillus subtilus cell free transcription-translation system for prototyping regulatory elements. Metab. Eng. 38, 370–381. doi: 10.1016/j.ymben.2016.09.008
Kelwick, R. J. R., Webb, A. J., and Freemont, P. S. (2020). Biological materials: the next frontier for cell-free synthetic biology. Front. Bioeng. Biotechnol. 8:399. doi: 10.3389/fbioe.2020.00399
Kenney, K. L., Smith, W. A., Gresham, G. L., and Westover, T. L. (2013). Understanding biomass feedstock variability. Biofuels 4, 111–127. doi: 10.4155/bfs.12.83
Khambhti, K., Hattacharjee, G., Gohil, N., Braddick, D., Kulkarni, V., and Singh, V. (2019). Exploring the potential of cell-free protein synthesis for extending the abilities of biological systems. Front. Bioeng. Biotechnol. 7:248. doi: 10.3389/fboie.2019.00248
Kopp, D., Willows, R. D., and Sunna, A. (2019a). Cell-free enzymatic conversion of spent coffee grounds into the platform chemical lactic acid. Front. Bioeng. Biotechnol. 7:389. doi: 10.3389/fbioe.2019.00389
Kopp, D., Willows, R. D., and Sunna, A. (2019b). Characterisation of the first archaeal mannonate dehydratase from Thermoplasma acidophilum and its potential in the catabolism of D-mannose. Catalysts 9:234. doi: 10.3390/catal9030234
Korman, T. P., Opgenorth, P. H., and Bowie, J. S. (2017). A synthetic biochemistry platform for cell free production of monoterpenes from glucose. Nat. Commun. 8:15526. doi: 10.1038/ncomms15526
Korman, T. P., Sahachartsiri, B., Li, D., Vinokur, J. M., Eisenberg, D., and Bowie, J. U. (2014). A synthetic biochemistry system for the in vitro production of isoprene from glycolysis intermediates. Protein Sci. 23, 576–585. doi: 10.1002/pro.2436
Koutinas, A. A., Vlysidis, A., Pleissner, D., Kopsahelis, N., Lopez Garcia, I., Kookos, I. K., et al. (2014). Valorization of industrial waste and byproduct streams via fermentation for the production of chemicals and biopolymers. Chem. Soc. Rev. 43, 2587–2627. doi: 10.1039/c3cs60293a
Krutsakorn, B., Honda, K., Ye, X., Imagawa, T., Bei, X., Okano, K., et al. (2013). In vitro production of n-butanol from glucose. Metab. Eng. 20, 84–91. doi: 10.1016/j.ymben.2013.09.006
Krüger, A., Mueller, A. P., Grant, A., Rybnicky, G. A., Engle, N. L., Zamin, K., et al. (2020). Development of a clostridia-based cell-free system for prototyping genetic parts and metabolic pathways. bioRxiv [Preprint]. doi: 10.1011/2020.03.11.987826
Lee, C. K., Monk, C. R., and Daniel, R. M. (2013). “Determination of enzyme thermal parameters for rational enzyme engineering and environmental/evolutionary studies,” in Protein Nanotechnology. Methods in Molecular Biology (Methods and Protocols), Vol. 996, ed. J. Gerrard (Totowa, NJ: Humana Press), 219–230. doi: 10.1007/978-1-62703-354-1_13
Li, Z., Li, F., Cai, L., Chen, Z., Qin, L., and Gao, X. D. (2020). One-pot multienzyme synthesis of rare ketoses from glycerol. J. Agric. Food Chem. 68, 1347–1353. doi: 10.1021/acs.jafc.9b06748
Li, Z., Yan, J., Sun, J., Xu, P., Ma, C., and Gao, C. (2018). Production of value-added chemicals from glycerol using in vitro enzymatic cascades. Commun. Chem. 1:71. doi: 10.1038/s42004-018-0070-7
Li, Z., Zhang, M., Jiang, T., Sheng, B., Ma, C., Xu, P., et al. (2017). Enzymatic cascades for efficient biotransformation of racemic lactate derived from corn steep water. ACS Sustainable Chem. Eng. 5, 3456–3464. doi: 10.1021/acssuschemeng.7b00136
Libicher, K., Hornberger, R., Heymann, M., and Mutschler, H. (2020). In vitro self-replication and multicistronic expression of large synthetic genomes. Nat. Commun. 11:904. doi: 10.1038/s41467-020-14694-2
Liu, Z., Zhang, Y., Jia, X., Hu, M., Deng, Z., Xu, Y., et al. (2017). In vitro reconstitution and optimization of the entire pathway to convert glucose into fatty acid. ACS Synth. Biol. 6, 701–709. doi: 10.1021/acssynbio.6b00348
López-Gallego, F. (2019). On-pot and cell-free biocatalysis using coimmobilized enzymes on advanced materials. Meth. Enzymol. 617, 385–411. doi: 10.1016/bs.mie.2018.12.013
Lu, F., Smith, P. R., Mehta, K., and Swartz, J. R. (2015). Development of a synthetic pathway to convert glucose to hydrogen using cell free extracts. Int. J. Hydrog. Energy 40, 9113–9124. doi: 10.1016/j.ijhydene.2015.05.121
Lu, Y. (2017). Cell-free synthetic biology: engineering in an open world. Synth. Syst. Biotechnol. 2, 23–27. doi: 10.1016/j.synbio.2017.02.003
Mahour, R., Klapproth, J., Rexer, T. F., Schildbach, A., Klamt, S., Pietzsch, M., et al. (2018). Establishment of a five-enzyme cell-free cascade for the synthesis of uridine diphosphate N-acetylglucosamine. J. Biotechnol. 283, 120–129. doi: 10.1016/j.jbiotec.2018.07.027
Marshall, R., Maxwell, C. S., Collins, S. P., Jacobsen, T., Luo, M. C., Begemann, M. B., et al. (2018). Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol. Cell 69, 146–157. doi: 10.1016/j.molcel.2017.12.007
Martín del Campo, J. S., Rollin, J., Myung, S., Chun, Y., Chandrayan, S., Patiño, R., et al. (2013). High-yield production of dihydrogen from xylose by using a synthetic enzyme cascade in a cell-free system. Angew. Chem. Int. Ed. Engl. 52, 4587–4590. doi: 10.1002/anie.201300766
Massaya, J., Prates Pereira, A., Mills-Lamptey, B., Benjamin, J., and Chuck, C. (2019). Conceptualization of a spent coffee grounds biorefinery: a review of existing valorisation approaches. Food Bioprod. Process. 118, 149–166. doi: 10.1016/j.fbp.2019.08.010
Meng, D., Wei, X., Zhang, Y.-H. P. J., Zhu, Z., You, C., and Ma, Y. (2018). Stoichiometric conversion of cellulosic biomass by in vitro Ssynthetic enzymatic biosystems for biomanufacturing. ACS Catal. 8, 9550–9559. doi: 10.1021/acscatal.8b02473
Moon, T. S., Yoon, S. H., Lanza, A. M., Roy-Mayhew, J. D., and Prather, K. L. T. (2009). Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Appl. Enviro. Microbiol. 75, 589–595. doi: 10.1128/AEM.01065-09
Moore, S. J., MacDonald, J. T., and Freemont, P. S. (2017a). Cell-free synthetic biology for in vitro prototype engineering. Biochem. Soc. Trans. 45, 785–791. doi: 10.1042/BST20170011
Moore, S. J., Tosi, T., Bell, D., Hleba, Y. B., Polizzi, K. M., and Freemont, P. S. (2017b). A cell-free synthetic biochemistry platform for raspberry ketone production. bioRxiv[Preprint]. doi: 10.1101/202341
Mukherjee, J., and Gupta, M. N. (2015). Biocatalysis for biomass valorization. Sustain. Chem. Process. 3, 7. doi: 10.1186/s40508-015-0037-2
Ninh, P. H., Honda, K., Sakai, T., Okano, K., and Ohtake, H. (2015). Assembly and multiple gene expression of thermophilic enzymes in Escherichia coli for in vitro metabolic engineering. Biotechnol. Bioeng. 112, 189–196. doi: 10.1002/bit.25338
Nirenberg, N. W., and Matthaei, J. H. (1961). The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. U.S.A 47, 1588–1602. doi: 10.1073/pnas.47.10.1588
Niu, F. X., Huang, Y. B., Shen, Y. P., Ji, L. N., and Liu, J. Z. (2020). Enhanced production of pinene by using a cell-free system with modular cocatalysis. J. Agri. Food Chem 68, 2139–2145. doi: 10.1021/acsjafc.9b07830
Nocek, B., Kochinyan, S., Proudfoot, M., Brown, G., Evdokimova, E., Osipiuk, J., et al. (2008). Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 17730–17735. doi: 10.1073/pnas.0807563105
Nowak, C., Beer, B., Pick, A., Roth, T., Lommes, P., and Sieber, V. (2015). A water-forming NADH oxidase from Lactobacillus pentosus suitable for the regeneration of synthetic biomimetic cofactors. Front. Microbiol. 6:957. doi: 10.3389/fmicb.2015.00957
Nowak, C., Pick, A., Lommes, P., and Sieber, V. (2017). Enzymatic reduction of nicotinamide biomimetic cofactors using an engineered glucose dehydrogenase: providing a regeneration system for artificial cofactors. ACS Catal. 7, 5202–5208. doi: 10.1021/acscatal.7b00721
Ogonah, O. W., Polizzi, K. M., and Bracewell, D. G. (2017). Cell free protein synthesis: a viable option for stratified medicines manufacturing. Curr. Opin. Biotechnol. 18, 77–83. doi: 10.1016/j.coche2017.10.003
Opgenorth, P. H., Korman, T. P., and Bowie, J. U. (2014). A synthetic biochemistry molecular purge valve module that maintains redox balance. Nat. Commun. 5, 1–8. doi: 10.1038/ncomms5113
Opgenorth, P. H., Korman, T. P., and Bowie, J. U. (2016). A synthetic biochemistry module for production of bio-based chemicals from glucose. Nat. Chem. Biol. 12, 393–395. doi: 10.1038/nchembio.2062
Pardee, K. (2018). Perspective: solidifying the impact of cell-free synthetic biology through lyophilization. Biochem. Eng. J. 138, 91–97.
Pardee, K., Slomovic, S., Nyugen, P. Q., Lee, J. W., Donghia, N., Burrill, D., et al. (2016). Portable, on-demand biomolecular manufacturing. Cell 167, 248–259. doi: 10.1016/cell.2016.09.013
Paul, C. E., and Hollmann, F. (2016). A survey of synthetic nicotinamide cofactors in enzymatic processes. Appl. Microbiol. Biotechnol. 100, 4773–4778. doi: 10.1007/s00253-016-7500-1
Peterson, M. E., Eisenthal, R., Danson, M. J., Spence, A., and Daniel, R. M. (2004). A new, intrinsic, thermal parameter for enzymes reveals true temperature optima. J. Biol. Chem. 279, 20717–20722. doi: 10.10-74/jbc.M309143200
Petroll, K., Care, A., Bergquist, P. L., and Sunna, A. (2020). A novel framework for the cell-free enzymatic production of glucaric acid. Metab. Eng. 57, 162–173. doi: 10.1016/j.ymben.2019.11.003
Petroll, K., Kopp, D., Care, A., Bergquist, P. L., and Sunna, A. (2019). Tools and strategies for constructing cell-free enzyme pathways. Biotechnol. Adv. 37, 91–108. doi: 10.1016/biotechadv.2018.11.007
Pleissner, D., and Lin, C. S. K. (2013). Valorisation of food waste in biotechnological processes. Sustain. Chem. Process. 1:21. doi: 10.1186/2043-7129-1-21
Poppe, L., and Vértessy, B. G. (2018). The fourth wave of biocatalysis emerges – The 13th International symposium on biocatalysis and biotransformations. Chembiochem 19, 284–287. doi: 10.1002/cbic.201700687
Ramalakshmi, K., Mohan, L. J., Takano-ishikawa, Y., and Goto, M. (2009). Bioactivities of low-grade green coffee and spent coffee in different in vitro model systems. Food Chem. 115, 79–85. doi: 10.1016/j.foodchem.2008.11.063
Reetz, M. T. (2013). Biocatalysis in organic chemistry and biotechnology: past, present and future. J. Amer. Chem. Soc. 135, 12480–12496. doi: 10.1021/ja405051fl
REN21 (2017). Renewable Energy Policy Network for the 21st Century. Renewables 2017 global status report. Available online at: (https://www.ren21.net/wp-content/uploads/2019/05/GSR2017_Full-Report_English.pdf) (accessed 1 April 2020).
Risso, V. A., Sanchez-Ruix, J. M., and Ozkan, S. B. (2018). Biotechnological and protein-engineering implications of ancestral protein resurrection. Curr. Opin. Struct. Biol. 51, 106–115. doi: 10.1016/j.sbi.208.02.007
Rolf, J., Rosenthal, K., and Lütz, S. (2019). Application of cell-free protein synthesis for faster biocatalyst development. Catalysts 9:190. doi: 10.3390/catal9020190
Rollin, J. A., Martin Del Campo, J., Myung, S., Sun, F., You, C., Bakovic, A., et al. (2015). High-yield hydrogen production from biomass by in vitro metabolic engineering: mixed sugars coutilization and kinetic modeling. Proc. Natl. Acad. Sci. U.S.A. 112, 4964–4969. doi: 10.1073/pnas.1417719112
Rosenblum, G., and Cooperman, B. S. (2014). Engine out of the chassis: cell-free protein synthesis and its uses. FEBS Lett. 588, 261–268. doi: 10.1016/febslet.2013
Rudroff, F., Mihovilovic, M. D., Gröger, H., Snajdrova, R., Idling, H., and Bornschuer, U. T. (2018). Opportunities and challenges for combining and chemo- and biocatalysis. Nat. Catal. 1, 12–22. doi: 10.1038/s41929-017-0010-4
Savile, C. K., Janey, J. M., Mungriff, E. C., Moore, J. C., Tam, S., Jarvis, W. R., et al. (2010). Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 329, 305–309. doi: 10.1126/science.1188934
Schillberg, S., Raven, N., Spiegel, H., Rasche, S., and Buntru, M. (2019). Critical analysis of the commercial potential of plants for the production of recombinant proteins. Front. Plant Sci. 10:720. doi: 10.3389/fpls.2019.00720
Schmidt-Dannert, C., and Lopez-Gallego, F. (2016). A roadmap for biocatalysis – functional and spatial orchestration of enzyme cascades. Microb. Biotechnol. 9, 601–609. doi: 10.1111/1751-7915.12386
Schoevaart, R., van Rantwijk, F., and Sheldon, R. A. (2000). A four-step enzymatic cascade for the one-pot synthesis of non-natural carbohydrates from glycerol. J. Org. Chem. 65, 6940–6943. doi: 10.1021/jo000492y
Schwander, T., Schada Von, Borzyskowski, L., Burgener, S., Cortina, N. S., and Erb, T. J. (2016). A synthetic pathway for the fixation of carbon dioxide in vitro. Science 354, 900–904. doi: 10.1126/science.aah5237
Shakeel, T., Sharma, A., and Yazdani, S. S. (2019). Building cell factories for the production of advanced fuels. Biochem. Soc. Trans. 47, 1701–1714. doi: 10.1042/BST20190168
Sheldon, R. A., and Brady, D. (2018). The limits to biocatalysis: pushing the envelope. Chem. Commun. 54, 6088–6104. doi: 10.1039/c8cc)2463d
Sheldon, R. A., and Woodley, J. M. (2018). Role of biocatalysis in sustainable chemistry. Chem. Rev. 118, 801–838.
Shi, T., Han, P., You, C., and Zhang, Y.-H. P. J. (2018). An in vitro synthetic biology platform for emerging industrial biomanufacturing: bottom-up pathway design. Synth. Syst. Biotechnol. 3, 186–195. doi: 10.1016/j.synbio.2018.05.002
Shimizu, Y., Inoue, A., Tomari, Y., Suzuki, T., Yokogawa, T., Nishikawa, K., et al. (2001). Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755. doi: 10.1038/90802
Shimizu, Y., Kanamori, T., and Ueda, T. (2005). Protein synthesis by pure translation systems. Methods 36, 299–304. doi: 10.1016/j.ymeth.2005.04.006
Silverman, A. D., Karim, A. S., and Jewett, M. C. (2020). Cell-free gene expression: an expanded repertoire of applications. Nat. Rev. Genet. 21, 151–170. doi: 10.1038/541576-019-0186-3
Smith, M., Wilding, K. M., Hunt, J. M., Bennett, A. M., and Bundy, B. C. (2014). The emerging age of cell-free synthetic biology. FEBS Lett. 588, 2755–2761. doi: 10.1016/febslet.2014.05.062
Sokic-Lazic, D., and Minteer, S. D. (2009). Pyruvate/air enzymatic biofuel cell capable of complete oxidation. Electrochem. Solid State Lett. 12, F26. doi: 10.1149/1.3170904
Sperl, J. M., and Sieber, V. (2018). Multienzyme cascade reactions-status and recent advances. ACS Catal. 8, 2385–2396. doi: 10.1021/acscatal.7b03440
Su, H. H., Guo, Z. W., Wu, X. L., Xu, P., Li, N., Zong, M. H., et al. (2019). Efficient bioconversion of sucrose to high−value−added glucaric acid by in vitro metabolic engineering. ChemSusChem. 12, 2278–2285. doi: 10.1002/cssc.201900185
Sun, S., Wei, X., and You, C. (2019). The construction of an in vitro synthetic enzymatic biosystem that facilitates laminaribiose biosynthesis from maltodextrin and glucose. Biotechnol. J. 14:1800493. doi: 10.1002/biot.201800493
Sutiono, S., Teshima, M., Beer, B., Schenk, G., and Sieber, V. (2020). Enabling the direct enzymatic dehydration of D-glycerate to pyruvate as the key step in synthetic enzyme cascades used in the cell-free production of fine chemicals. ACS Catal. 10, 3110–3118. doi: 10.1021/acscatal.9b05068
Swartz, J. (2006). Developing cell-free biology for industrial application. J. Ind. Microbiol. Biotechnol. 33, 476–485. doi: 10.1007/s10295-006-0127-y
Swartz, J. R. (2012). Transforming biochemical engineering with cell-free biology. AICHE J. 58, 5–3. doi: 10.1002/aic.1370
Swartz, J. R. (2018). Expanding biological applications using cell-free metabolic engineering. Metab. Eng. 50, 156–172. doi: 10.1016/j.ymben.2018.09.011
Takao, I., Fujii, S., Ishii, A., Han, L., Kumao, T., Ozaki, K., et al. (2006). Effects of mannooligosaccharides from coffee mannan on fat storage in mice fed a high fat diet. J. Heal. Sci. 52, 333–337. doi: 10.1248/jhs.52.333
Taniguchi, H., Okano, K., and Honda, K. (2017). Modules for in vitro metabolic engineering: pathway assembly for bio-based production of value-added chemicals. Synth. Syst. Biotechnol. 2, 65–72. doi: 10.1016/.synbio.2017.06.00
Thaore, V. B., Armstrong, R. D., Hutchings, G. J., Knight, D. W., Chadwick, D., and Shah, N. (2020). Sustainable production of glucaric acid from corn stover via glucose oxidation: an assessment of homogeneous and heterogeneous catalytic oxidation production routes. Chem. Eng. Res. Des. 153, 337–349. doi: 10.1016/j.cherd.2019.10.042
Tian, C., Yang, J., Zeng, Y., Zhang, T., Zhou, Y., Men, Y., et al. (2019). Biosynthesis of raffinose and stachyose from sucrose via an in vitro multienzyme system. Appl. Environ. Microbiol. 85:e02306-18. doi: 10.1128/AEM.02306-18
Tinafar, A., Jaenes, K., and Pardee, K. (2019). Synthetic biology goes cell-free. BMC Biol. 17:64. doi: 10.1186/s12915-019-0685-x
Torres, P., and Batista-Viera, F. (2019). Production of D-tagatose and d-fructose from whey by co-immobilized enzymatic system. Mol. Catal. 463, 99–109. doi: 10.1016/j.mcat.2018.11.017
Tuck, C. O., Pérez, E., Horváth, I. T., Sheldon, R. A., and Poliakoff, M. (2012). Valorization of biomass: Deriving more value from waste. Science 337, 695–699. doi: 10.1126/science.1218930
United Nations Environment Programme [UNEP] (2015). Programme Performance Report 2015. Available online at: https://www.unenvironment.org/annualreport/2015. (accessed 10 April 2020)
United Nations Environment Programme [UNEP] (2018). Programme Performance Report 2018. Available online at: https://www.unenvironment.org/annualreport/2018. (Accessed 10 April 2020)
US Department of Energy, Office of Energy Efficiency & Renewable Energy. (2018). Cell-Free Synthetic Biology and Biocatalysis: Prototyping and Conversion Platforms. Washington, DC: US Department of Energy, Office of Energy Efficiency & Renewable Energy.
Van der Donk, W. A., and Zhao, H. (2003). Recent developments in pyridine nucleotide regeneration. Curr. Opin. Biotechnol. 14, 421–426. doi: 10.1016/S0958-1669(03)00094-6
Vardon, D. R., Moser, B. R., Zheng, W., Witkin, K., Evangelista, R. L., Strathmann, T. J., et al. (2013). Complete utilization of spent coffee grounds to produce biodiesel, bio-oil, and biochar. ACS Sustain. Chem. Eng. 1, 1286–1294. doi: 10.1021/sc400145w
Velasco-Lozano, S., Benítez-Mateos, A. I., and López-Gallego, F. (2017). Co-immobilized phosphorylated cofactors and enzymes as self-sufficient heterogeneous biocatalysts for chemical processes. Angew. Chem. Int. Ed. Engl. 56, 771–775. doi: 10.1002/anie.201609758
Voloshin, A. M., and Swartz, J. R. (2005). Efficient and scalable method for scaling up cell free protein synthesis in batch mode. Biotechnol. Bioeng. 91, 516–521. doi: 10.1002/bit.20528
Wang, W., Liu, M., You, C., Li, Z., and Zhang, Y.-H. P. (2017). ATP-free biosynthesis of a high-energy phosphate metabolite fructose 1,6-diphosphate by in vitro metabolic engineering. Metab. Eng. 42, 168–174. doi: 10.10-16/j.ymben.2017.06.006
Welch, P., and Scopes, R. K. (1985). Studies on cell-free metabolism: ethanol production by a yeast glycolytic system reconstituted from purified enzymes. J. Biotechnol. 2, 257–273. doi: 10.1016/0168-1656(85)90029-X
Whittaker, J. W. (2013). Cell-free protein synthesis: the state of the art. Biotechnol. Lett. 35, 143–152. doi: 10.1007/s10529-012-1075-40
Wichelecki, D. J. (2018). Enzymatic Synthesis of D-tagatose. U.S. Patent 20180216146. Geneva: World Intellectual Property Organization.
Wilding, K. M., Schinn, S. M., Long, E. A., and Bundy, B. C. (2018). The emerging impact of cell-free chemical biosynthesis. Curr. Opin. Biotechnol. 53, 115–121. doi: 10.1016/1288j.copbio.2017.12019
Woodward, J., Orr, M., Cordray, K., and Greenbaum, E. (2000). Biotechnology: enzymatic production of biohydrogen. Nature 405, 1014–1015. doi: 10.1038/35016633
Wu, Y., Mao, G., Fan, H., Song, A., Zhang, Y. P., and Chen, H. (2017). Biochemical properties of GH94 cellodextrin phosphorylase THA_1941 from a thermophilic eubacterium Thermosipho africanus TCF52B with cellobiose phosphorylase activity. Sci. Rep. 7:4849. doi: 10.1038/s41598-017-05289-x
Xie, L., Wei, X., Zhou, X., Meng, D., Zhou, R., Zhang, Y.-H. P., et al. (2018). Conversion of D-glucose to L-lactate via pyruvate by an optimized cell-free enzymatic biosystem containing minimized reactions. Synth. Syst. Biotechnol. 3, 204–210. doi: 10.1016/j.synbio.2018.05.003
Xu, S., and Minteer, S. D. (2012). Enzymatic biofuel cell for oxidation of glucose to CO2. ACS Catal. 2, 91–94. doi: 10.1021/cs200523s
Yan, N., and Chen, X. (2015). Sustainability: don’t waste seafood waste. Nature 524, 155–157. doi: 10.1038/524155a
Yang, J., Zhu, Y., Qu, G., Zeng, Y., Tian, C., Dong, C., et al. (2018). Biosynthesis of dendroketose from different carbon sources using in vitro and in vivo metabolic engineering strategies. Biotechnol. Biofuels 11:290. doi: 10.1186/s13068-018-1293-7
Yang, W. C., Patel, K. G., Wong, H. E., and Swartz, J. R. (2012). Simplifying and streamlining Escherichia coli-based cell-free protein synthesis. Biotechnol. Prog. 28, 413–420. doi: 10.1002/btpro.1059
Ye, X., Honda, K., Morimoto, Y., Okano, K., and Ohtake, H. (2013). Direct conversion of glucose to malate by synthetic metabolic engineering. J. Biotechnol. 164, 34–40. doi: 10.1016/j.biotec.2012.11.011
Ye, X., Honda, K., Sakai, T., Okano, K., Omasa, T., Hirota, R., et al. (2012). Synthetic metabolic engineering-a novel, simple technology for designing a chimeric metabolic pathway. Microb. Cell Factories 11:120. doi: 10.1186/1475-2859-11-120
Ye, X., Wang, Y., Hopkins, R. C., Adams, M. W., Evans, B. R., Mielenz, J. R., et al. (2009). Spontaneous high-yield production of hydrogen from cellulosic materials and water catalyzed by enzyme cocktails. ChemSusChem 2, 149–152. doi: 10.1002/cssc.200900017
Yim, H., Haselbeck, R., Niu, W., Pujol-Baxley, C., Burgard, A., Boldt, J., et al. (2011). Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 7, 445–452. doi: 10.1038/nchembio.580
You, C., Shi, T., Li, Y., Han, P., Zhou, X., and Zhang, Y.-H. P. (2017). An in vitro synthetic biology platform for the industrial biomanufacturing of myo−inositol from starch. Biotechnol. Bioeng. 114, 1855–1864. doi: 10.1002/bit.26314
Yu, X., Liu, T., Zhu, F., and Khosla, C. (2011). In vitro reconstitution and steady-state analysis of the fatty acid synthase from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 108, 18643–18648. doi: 10.1073/pnas.1110852108
Yu, Y., Hu, C., Xia, L., and Wang, J. (2018). Artificial metalloenzyme design with unnatural amino acids and non-native cofactors. ACS Catal. 2018, 1851–1863. doi: 10.1021/acscatal.7b03754
Zawada, J. F., Yin, G., Steiner, A. R., Yang, J., Naresh, A., Sushmita, M. R., et al. (2011). Microscale manufacturing scale-up of cell-free cytokine production-a new approach for shortening protein production development timelines. Biotechnol. Bioeng. 108, 1570–1578. doi: 10.1002/bit.23103
Zhang, Y.-H. P., Sun, J., and Ma, Y. (2017). Biomanufacturing: history and perspective. J. Ind. Microbiol. Biotechnol. 44, 773–784. doi: 10.1007/s10295-016-1863-2
Zhao, H., and van der Donk, W. A. (2003). Regeneration of cofactors for use in biocatalysis. Curr. Opin. Biotechnol. 14, 583–589. doi: 10.1007/s12551-020-00618-3
Zhong, C., and Nidetzky, B. (2020). Three−enzyme phosphorylase cascade for integrated production of short−chain cellodextrins. Biotechnol. J. 15:1900349. doi: 10.1002/biot.201900349
Zhong, C., Wei, P., and Zhang, Y.-H. P. (2017a). A kinetic model of one-pot rapid biotransformation of cellobiose from sucrose catalyzed by three thermophilic enzymes. Chem. Eng. Sci. 161, 159–166. doi: 10.1016/j.ces.2016.11.047
Zhong, C., You, C., Wei, P., and Zhang, Y.-H. P. (2017b). Thermal cycling cascade biocatalysis of myo-inositol synthesis from sucrose. ACS Catal. 7, 5992–5999. doi: 10.1021/acscatal.7b01929
Zhou, H. Y., Yong, J., Gao, H., Li, T., Xiao, H. S., and Wu, Y. Y. (2017). Mannanase Man23 mutant library construction based on a novel protein expression system. J. Sci. Food Agric. 97, 2199–2204. doi: 10.1002/jsfa.8029
Zhou, Z., and Roelfes, G. (2020). Synergistic catalysis in an artificial enzyme by simultaneous action of two abiological catalytic sites. Nat. Catal. 3, 289–294. doi: 10.1038/s41929-019-0420-6
Zhu, Z., and Zhang, Y.-H. P. (2017). In vitro metabolic engineering of bioelectricity generation by the complete oxidation of glucose. Metab. Eng. 39, 110–116. doi: 10.1016/j.ymben.2016.11.002
Zubay, G. (1973). In vitro synthesis of protein in microbial systems. Ann. Rev. Genet. 7, 267–287. doi: 10.1146/annurev.ge.07.120173.00141
Keywords: cell-free biocatalysis, cell-free synthesis, multi-enzyme cascades, in vitro biocatalysis, synthetic biology, pathway design
Citation: Bergquist PL, Siddiqui S and Sunna A (2020) Cell-Free Biocatalysis for the Production of Platform Chemicals. Front. Energy Res. 8:193. doi: 10.3389/fenrg.2020.00193
Received: 15 June 2020; Accepted: 20 July 2020;
Published: 27 August 2020.
Edited by:
Michael C. Jewett, Northwestern University, United StatesReviewed by:
Richard Kelwick, Imperial College London, United KingdomChun You, Tianjin Institute of Industrial Biotechnology (CAS), China
Copyright © 2020 Bergquist, Siddiqui and Sunna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anwar Sunna, anwar.sunna@mq.edu.au
 Peter L. Bergquist
Peter L. Bergquist Sana Siddiqui
Sana Siddiqui Anwar Sunna
Anwar Sunna