Abstract
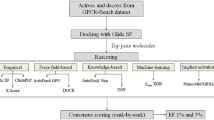
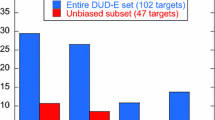
Recent breakthroughs in G protein-coupled receptor (GPCR) crystallography and the subsequent increase in number of solved GPCR structures has allowed for the unprecedented opportunity to utilize their experimental structures for structure-based drug discovery applications. As virtual screening represents one of the primary computational methods used for the discovery of novel leads, the GPCR-Bench dataset was created to facilitate comparison among various virtual screening protocols. In this study, we have benchmarked the performance of Molecular Mechanics/Poisson-Boltzmann Surface Area (MM/PBSA) in improving virtual screening enrichment in comparison to docking with Glide, using the entire GPCR-Bench dataset of 24 GPCR targets and 254,646 actives and decoys. Reranking the top 10% of the docked dataset using MM/PBSA resulted in improvements for six targets at EF1% and nine targets at EF5%, with the gains in enrichment being more pronounced at the EF1% level. We additionally assessed the utility of rescoring the top ten poses from docking and the ability of short MD simulations to refine the binding poses prior to MM/PBSA calculations. There was no clear trend of the benefit observed in both cases, suggesting that utilizing a single energy minimized structure for MM/PBSA calculations may be the most computationally efficient approach in virtual screening. Overall, the performance of MM/PBSA rescoring in improving virtual screening enrichment obtained from docking of the GPCR-Bench dataset was found to be relatively modest and target-specific, highlighting the need for validation of MM/PBSA-based protocols prior to prospective use.



Similar content being viewed by others
Abbreviations
- 5HT1B:
-
5-Hydroxytryptamine 1B receptor
- 5HT2B:
-
5-Hydroxytryptamine 2B receptor
- AA2AR:
-
Adenosine A2A receptor
- ACM2:
-
Muscarinic acetylcholine 2 receptor
- ACM3:
-
Muscarinic acetylcholine 3 receptor
- ADRB1:
-
Beta-1 adrenergic receptor
- ADRB2:
-
Beta-2 adrenergic receptor
- BEAR:
-
Binding Estimation After Refinement
- CCR5:
-
C-C chemokine receptor type 5
- CRFR1:
-
Corticotropin releasing factor receptor 1
- CXCR4:
-
C-X-C chemokine receptor type 4
- DRD3:
-
Dopamine 3 receptor
- GPCR:
-
G protein-coupled receptor
- GPR40:
-
Free fatty acid receptor 1
- EF:
-
Enrichment factor
- HRH1:
-
Histamine 1 receptor
- MD:
-
Molecular dynamics
- MGLUR1:
-
Metabotropic glutamate receptor 1
- MGLUR5:
-
Metabotropic glutamate receptor 5
- MM/GBSA:
-
Molecular Mechanics/Generalized Born Surface Area
- MM/PBSA:
-
Molecular Mechanics/Poisson-Boltzmann Surface Area
- OPRD:
-
Delta opioid receptor
- OPRK:
-
Kappa opioid receptor
- OPRM:
-
Mu opioid receptor
- OPRX:
-
Nociceptin receptor
- OX2R:
-
Orexin receptor type 2
- PAR1:
-
Proteinase-activated receptor 1
- P2Y12:
-
P2Y purinoceptor 12
- S1PR1:
-
Sphingosine 1-phosphate receptor 1
- SMO:
-
Smoothened receptor
- TM:
-
Transmembrane
References
Fredriksson R, Lagerström MC, Lundin L-G, Schiöth HB (2003) The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol 63:1256–1272. https://doi.org/10.1124/mol.63.6.1256
Kolakowski LF (1994) GCRDb: a G-protein-coupled receptor database. Receptors Channels 2:1–7
Lagerstrom MC, Schioth HB (2008) Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov 7:339–357. https://doi.org/10.1038/nrd2518
Katritch V, Cherezov V, Stevens RC (2013) Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol 53:531–556. https://doi.org/10.1146/annurev-pharmtox-032112-135923.Structure-Function
Sriram K, Insel PA (2018) GPCRs as targets for approved drugs: How many targets and how many drugs? Mol Pharmacol. https://doi.org/10.1124/mol.117.111062
Ghosh E, Kumari P, Jaiman D, Shukla AK (2015) Methodological advances: the unsung heroes of the GPCR structural revolution. Nat Rev Mol Cell Biol 16:69–81. https://doi.org/10.1038/nrm3933
Pándy-Szekeres G, Munk C, Tsonkov TM et al (2018) GPCRdb in 2018: Adding GPCR structure models and ligands. Nucleic Acids Res 46:D440–D446. https://doi.org/10.1093/nar/gkx1109
Michino M, Abola E, Brooks CL et al (2009) Community-wide assessment of GPCR structure modelling and ligand docking: GPCR Dock 2008. Nat Rev Drug Discov 8:455–463. https://doi.org/10.1038/nrd2877
Beuming T, Sherman W (2012) Current assessment of docking into GPCR crystal structures and homology models: Successes, challenges, and guidelines. J Chem Inf Model 52:3263–3277. https://doi.org/10.1021/ci300411b
Loo JSE, Emtage AL, Ng KW et al (2018) Assessing GPCR homology models constructed from templates of various transmembrane sequence identities: Binding mode prediction and docking enrichment. J Mol Graph Model 80:38–47. https://doi.org/10.1016/j.jmgm.2017.12.017
Cheng T, Li X, Li Y et al (2009) Comparative assessment of scoring functions on a diverse test set. J Chem Inf Model 49:1079–1093. https://doi.org/10.1021/ci9000053
Plewczynski D, Lazniewski M, Augustyniak R, Ginalski K (2011) Can we trust docking results? Evaluation of seven commonly used programs on PDBbind database. J Comput Chem 32:742–755. https://doi.org/10.1002/jcc
Wang Z, Sun H, Yao X et al (2016) Comprehensive evaluation of ten docking programs on a diverse set of protein–ligand complexes: the prediction accuracy of sampling power and scoring power. Phys Chem Chem Phys 18:12964–12975. https://doi.org/10.1039/C6CP01555G
Kollman PA, Massova I, Reyes C et al (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897. https://doi.org/10.1021/ar000033
Adcock SA, McCammon JA (2006) Molecular dynamics: Survey of methods for simulating the activity of proteins. Chem Rev 106:1589–1615. https://doi.org/10.1021/cr040426m
Kim JT, Hamilton AD, Bailey CM et al (2006) FEP-guided selection of bicyclic heterocycles in lead optimization for non-nucleoside inhibitors of HIV-1 reverse transcriptase. J Am Chem Soc 128:15372–15373. https://doi.org/10.1021/ja076881s
Laio A, Parrinello M (2002) Escaping free-energy minima. Proc Natl Acad Sci U S A 99:12562–12566. https://doi.org/10.1073/pnas.202427399
Singh N, Warshel A (2010) Absolute binding free energy calculations: on the accuracy of computational scoring of protein-ligand interactions. Proteins 78:1705–1723. https://doi.org/10.1002/prot.22687
Sun H, Li Y, Shen M et al (2014) Assessing the performance of MM/PBSA and MM/GBSA methods. 5. Improved docking performance using high solute dielectric constant MM/GBSA and MM/PBSA rescoring. Phys Chem Chem Phys 16:22035–22045. https://doi.org/10.1039/c4cp03179b
Hou T, Wang J, Li Y, Wang W (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods: I. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51:69–82. https://doi.org/10.1021/ci100275a.Assessing
Steinbrecher T, Case DA, Labahn A (2006) A multistep approach to structure-based drug design: studying ligand binding at the human neutrophil elastase. J Med Chem 49:1837–1844. https://doi.org/10.1021/jm0505720
Thompson DC, Humblet C, Joseph-McCarthy D (2008) Investigation of MM-PBSA rescoring of docking poses. J Chem Inf Model 48:1081–1091. https://doi.org/10.1021/ci700470c
Anighoro A, Rastelli G (2013) Enrichment factor analyses on G-protein coupled receptors with known crystal structure. J Chem Inf Model 53:739–743. https://doi.org/10.1021/ci4000745
Degliesposti G, Portioli C, Parenti MD, Rastelli G (2011) BEAR, a novel virtual screening methodology for drug discovery. J Biomol Screen 16:129–133. https://doi.org/10.1177/1087057110388276
Ferrari AM, Degliesposti G, Sgobba M, Rastelli G (2007) Validation of an automated procedure for the prediction of relative free energies of binding on a set of aldose reductase inhibitors. Bioorgq Med Chem 15:7865–7877. https://doi.org/10.1016/j.bmc.2007.08.019
Rastelli G, Degliesposti G, Del Rio A, Sgobba M (2009) Binding estimation after refinement, a new automated procedure for the refinement and rescoring of docked ligands in virtual screening. Chem Biol Drug Des 73:283–286. https://doi.org/10.1111/j.1747-0285.2009.00780.x
Rastelli G, Del RA, Degliesposti G, Sgobba M (2010) Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J Comput Chem 31:797–810. https://doi.org/10.1002/jcc.21372
Virtanen SI, Niinivehmas SP, Pentikäinen OT (2015) Case-specific performance of MM-PBSA, MM-GBSA, and SIE in virtual screening. J Mol Graph Model 62:303–318. https://doi.org/10.1016/j.jmgm.2015.10.012
Congreve M, Langmead CJ, Mason JS, Marshall FH (2011) Progress in structure based drug design for G protein-coupled receptors. J Med Chem 54:4283–4311. https://doi.org/10.1021/jm200371q
Shoichet BK, Kobilka BK (2012) Structure-based drug screening for G-protein-coupled receptors. Trends Pharmacol Sci 33:268–272. https://doi.org/10.1016/j.tips.2012.03.007
Weiss DR, Bortolato A, Tehan B, Mason JS (2016) GPCR-Bench: a benchmarking set and practitioners’ guide for G protein-coupled receptor docking. J Chem Inf Model 56:642–651. https://doi.org/10.1021/acs.jcim.5b00660
Mysinger MM, Carchia M, Irwin JJ, Shoichet BK (2012) Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J Med Chem 55:6582–6594. https://doi.org/10.1021/jm300687e
Dassault Systèmes BIOVIA (2015) Pipeline Pilot. Dassault Systèmes, San Diego
Milletti F, Storchi L, Sforna G, Cruciani G (2007) New and original pKa prediction method using grid molecular interaction fields. J Chem Inf Model 47:2172–2181. https://doi.org/10.1021/ci700018y
Milletti F, Storchi L, Sfoma G et al (2009) Tautomer enumeration and stability prediction for virtual screening on large chemical databases. J Chem Inf Model 49:68–75. https://doi.org/10.1021/ci800340j
Molecular Networks GmbH (2011) 3D Structure Generator CORINA Classic. Molecular Networks GmbH, Nürnberg
Friesner RA, Banks JL, Murphy RB et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking acuracy. J Med Chem 47:1739–1749. https://doi.org/10.1021/jm0306430
Halgren TA, Murphy RB, Friesner RA et al (2004) Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 47:1750–1759. https://doi.org/10.1021/jm030644s
Glide (2019) Schrödinger, LLC, New York
Abraham MJ, Hess B, van der Spoel D, Lindahl E (2018) GROMACS User Manual version 2018
Duan Y, Wu C, Chowdhury S et al (2003) A point-charge force field for molecular mechanics simulations of proteins. J Comput Chem 24:1999
Wang J, Wolf RM, Caldwell JW et al (2004) Development and testing of a general Amber force field. J Comput Chem 25:1157–1174. https://doi.org/10.1002/jcc.20035
Jakalian A, Jack DB, Bayly CI (2002) Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem 23:1623–1641. https://doi.org/10.1002/jcc.10128
Sousa da Silva AW, Vranken WF (2012) ACPYPE - AnteChamber PYthon Parser interfacE. BMC Res Notes 5:367. https://doi.org/10.1186/1756-0500-5-367
Wennberg CL, Murtola T, Hess B, Lindahl E (2013) Lennard-Jones lattice summation in bilayer simulations has critical effects on surface tension and lipid properties. J Chem Theory Comput 9:3527–3537. https://doi.org/10.1021/ct400140n
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092. https://doi.org/10.1063/1.464397
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472. https://doi.org/10.1002/(SICI)1096-987X(199709)18:12<1463:AID-JCC4>3.0.CO;2-H
Hess B (2008) P-LINCS: a parallel linear constraint solver for molecular simulation. J Chem Theory Comput 4:116–122. https://doi.org/10.1021/ct700200b
Jorgensen WL, Chandrasekhar J, Madura JD et al (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935. https://doi.org/10.1063/1.445869
Nosé S, Klein ML (1983) Constant pressure molecular dynamics for molecular systems. Mol Phys 50:1055–1076. https://doi.org/10.1080/00268978300102851
Evans DJ, Holian BL (1985) The Nose–Hoover thermostat. J Chem Phys 83:4069–4074. https://doi.org/10.1063/1.449071
Kumari R, Kumar R, Lynn A (2014) g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54:1951–1962. https://doi.org/10.1021/ci500020m
Sgobba M, Caporuscio F, Anighoro A et al (2012) Application of a post-docking procedure based on MM-PBSA and MM-GBSA on single and multiple protein conformations. Eur J Med Chem 58:431–440. https://doi.org/10.1016/j.ejmech.2012.10.024
Hou T, Wang J, Li Y, Wang W (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods: II. The accuracy of ranking poses generated from docking. J Comput Chem 32:866–877. https://doi.org/10.1002/jcc.21666
Sun H, Li Y, Shen M et al (2014) Assessing the performance of MM/PBSA and MM/GBSA methods. 5. Improved docking performance by using high solute dielectric constant MM/GBSA and MM/PBSA rescoring. Phys Chem Chem Phys 16:22035–22045. https://doi.org/10.1039/C4CP03179B
El Khoury L, Santos-Martins D, Sasmal S et al (2019) Comparison of affinity ranking using AutoDock-GPU and MM-GBSA scores for BACE-1 inhibitors in the D3R Grand Challenge 4. J Comput Aided Mol Des. https://doi.org/10.1007/s10822-019-00240-w
Oehme DP, Brownlee RTC, Wilson DJD (2012) Effect of atomic charge, solvation, entropy, and ligand protonation state on MM-PB(GB)SA binding energies of HIV Protease. J Comput Chem 33:2566–2580. https://doi.org/10.1002/jcc.23095
Sun H, Li Y, Tian S et al (2014) Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys Chem Chem Phys 16:16719. https://doi.org/10.1039/C4CP01388C
Wang C, Nguyen PH, Pham K et al (2016) Calculating protein–ligand binding affinities with MMPBSA: method and error analysis. J Comput Chem 37:2436–2446. https://doi.org/10.1002/jcc.24467
Yang T, Wu JC, Yan C et al (2011) Virtual screening using molecular simulations. Proteins 79:1940–1951. https://doi.org/10.1002/prot.23018
Ramírez D, Caballero J (2018) Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules 23:1–17. https://doi.org/10.3390/molecules23051038
Su M, Du Y, Yang Q et al (2018) Comparative assessment of scoring functions: the CASF-2016 update. J Chem Inf Model. https://doi.org/10.1021/acs.jcim.8b00545
Yau MQ, Emtage AL, Chan NJY et al (2019) Evaluating the performance of MM/PBSA for binding affinity prediction using class A GPCR crystal structures. J Comput Aided Mol Des 33:487–496. https://doi.org/10.1007/s10822-019-00201-3
Kuhn B, Gerber P, Schulz-Gasch T, Stahl M (2005) Validation and use of the MM-PBSA approach for drug discovery. J Med Chem 48:4040–4048. https://doi.org/10.1021/jm049081q
Greenidge PA, Kramer C, Mozziconacci JC, Wolf RM (2013) MM/GBSA binding energy prediction on the PDBbind data set: successes, failures, and directions for further improvement. J Chem Inf Model 53:201–209. https://doi.org/10.1021/ci300425v
Funding
This research was supported by Taylor’s University through its Taylor’s University Flagship Research Grant Scheme under grant number TUFR/2017/002/10 and Taylor’s PhD Scholarship Program.
Author information
Authors and Affiliations
Contributions
All authors gave approval to the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yau, M.Q., Emtage, A.L. & Loo, J.S.E. Benchmarking the performance of MM/PBSA in virtual screening enrichment using the GPCR-Bench dataset. J Comput Aided Mol Des 34, 1133–1145 (2020). https://doi.org/10.1007/s10822-020-00339-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-020-00339-5