Abstract
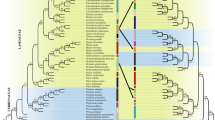
Taxonomic complexities, like environmental plasticity and homoplasy, make precise identification challenging in Calamus, the genus of spiny climbing palms of the subfamily Calamoideae (Arecaceae). In the present study, the species discriminatory power of twelve potential DNA barcode regions (rbcL, matK, psbA-trnH, rpoC, rpoB, psbK-psbI, atpF-atpH, psbZ-trnfM, ITS1, ITS2, PRK, and RPB2) were evaluated in 21 species of Calamus from the Western Ghats region of India, using distance, tree, and similarity based statistical methods. Except for the low copy nuclear region, RPB2, none of the tested plastid loci or nuclear loci ITS, either singly or in combinations, could discriminate all the species of Calamus due to low substitution rate of plastid regions and multiple copies of ITS respectively. The RPB2 locus showed highest species resolution with 96% accuracy in similarity based analysis, indicating its potential and efficiency as a barcode locus for the genus. The putative “Calamus gamblei complex” based on overlapping morphology was successfully resolved as six distinct, though closely related, species. The analysis also indicates that C. delessertianus is a morphological variant of C. dransfieldii. In spite of being a low copy nuclear gene region, RPB2 provided an efficient barcode to delineate Calamus species and has the potential to further extend its use as a prospective barcode to other Palm genera.



Similar content being viewed by others
Data Availability Statement
All the nucleotide sequence data generated in the study was submitted to NCBI and the list of accession numbers are provided as supplementary table S1.
References
Ahmad I, Khan S, Naeem M, Hayat M, Azmi UR, Ahmed S, Murtaza G, Irfan M (2019) Molecular identification of ten palm species using DNA Fingerprinting. Int J Pure App Biosci 7(1):46–51
Al-Qurainy F, Khan S, Al-Hemaid FM, Ajmal Ali A, Tarroum M, Ashraf M (2011) Assessing molecular signature for some potential date (Phoenix dactylifera L.) cultivars from Saudi Arabia, based on chloroplast DNA sequences rpoB and psbA trnH. Int J Mol Sci 12:6871–6880
Asif MJ, Cannon CH (2005) DNA extraction from processed wood: a case study for the identification of an endangered timber species (Gonystylus bancanus). Plant Mol Biol Rep 23:185–192
Atria M, Mil H, Baker WJ, Dransfield J, Welzen PC (2017) Morphometric analysis of rattan Calamus javensis complex (Arecaceae:Calamoideae). Syst Bot 42:494–506
Baker WJ, Hedderson TA, Dransfield J (2000) Molecular phylogenetics of subfamily Calamoideae (Palmae) based on nrDNA ITS and cpDNA rps16 intron sequence data. Mol Phylogenet Evol 14:195–217
Baker DA, Stevenson DW, Little DP (2012) DNA barcode identification of black cohosh herbal dietary supplements. J AOAC Int 95(4):1023–1034
Ballardini M, Mercuri A, Littardi C et al (2013) The chloroplast DNA locus psbZ-trnfM as a potential barcode marker in Phoenix L. (Arecaceae). ZooKeys 365:71–82
Bayton RP (2005) Borassus L. and the Borassoid palms: systematics and evolution. Ph.D. Thesis, University of Reading, UK
Beccari O (1908) Asiatic palms—Lepidocaryeae. Part II. The species of Calamus. Ann R Bot Gard 11:1–518
Boer WJ (1968) The Genomoid palms. Verh Kon Ned Akad Wetensch Afd Natuurk Tweede Sect Ser 58:1–202
CBOL Plant Working Group (2009) A DNA barcode for land plants. Proc Natl Acad Sci USA 106:12794–12797
Chase MW, Salamin N, Wilkinson M et al (2005) Land plants and DNA barcodes: short-term and long-term goals. Philos Trans R Soc Lond B Biol Sci 360:1889–1895
Chen S, Yao H, Han J et al (2010) Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 5:e8613
China Plant BOL Group, Li DZ, Gao LM, Li HT et al (2011) Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci USA 108:19641–19646
Daru BH, van der Bank M, Bello A, Yessoufou K (2017) Testing the reliability of standard and complementary DNA barcodes for the monocot subfamily Alooideae from South Africa. Genome 60:337–347
Denton AL, Mc Conaughy BL, Hall BD (1998) Usefulness of RNA polymerase II coding sequences for estimation of green plant phylogeny. Mol Biol Evol 15:1082–1085
Dev SA, Muralidharan EM, Sujanapal P, Balasundaran M (2014) Identification of market adulterants in East Indian sandalwood using DNA barcoding. Ann For Sci 71:517–522
Dev SA, Sijimol K, Prathibha PS, Sreekumar VB, Muralidharan EM (2020) DNA barcoding as a valuable molecular tool for the certification of planting materials in bamboo. 3 Biotech 10:59
Doyle JJ, Doyle JL (1990) A rapid total DNA preparation procedure for fresh plant tissue. Focus 12:13–15
Dransfield J (1992) The rattans of Sarawak. Royal Botanic Gardens and Sarawak Forest Department, Kew
Dransfield J, Manokaran N (1994) Rattans. Plant Resources of South-East Asia No. 6. Indonesia, PROSEA Foundation, Bogor
Duan H, Wang W, Zeng Y, Guo M, Zhou YI (2019) The screening and identification of DNA barcode sequences for Rehmannia. Sci Rep 9 (1):17295
Edwards D, Horn A, Taylor D, Savolainen V, Hawkins JA (2008) DNA barcoding of a large genus, Aspalathus L. (Fabaceae). Taxon 57:1317–1327
Elansary HO, Ashfaq M, Ali HM, Yessoufou K (2017) The first initiative of DNA barcoding of ornamental plants from Egypt and potential applications in horticulture industry. PLoS ONE 12(2):e0172170
Enan MR, Ahmed A (2016) Cultivar-level phylogeny using chloroplast DNA barcode psbK-psbI spacers for identification of Emirati date palm (Phoenix dactylifera L.) varieties. Genet Mol Res 15:3. https://doi.org/10.4238/gmr.15038470
Erickson DL, Spouge J, Resch A, Weigt LA, Kress JW (2008) DNA barcoding in land plants: developing standards to quantify and maximize success. Taxon 57:1304–1316
Feng S, Jiang Y, Wang S et al (2013) Molecular identification of Dendrobium Species (Orchidaceae) based on the DNA barcode ITS2 region and its application for phylogenetic study. Int J Mol Sci 16:21975–21988
Ford CS, Ayres KL, Toomey N et al (2009) Selection of candidate coding DNA barcoding regions for use on land plants. Bot J Linn Soc 159:1–11
Fuji T (2007) Outline of the research project “Methods to identify wood species and origin of timber of Southeast Asia”. In: Proceedings of the international symposium on development of improved methods to identify Shorea species wood and its origin, September 25–26, 2007. University of Tokyo, Tokyo and IUFRO, 19
Gao T, Yao H, Song JY et al (2010) Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J Ethnopharmacol 130:116–121
Gaut BS, Muse SV, Clark WD, Clegg MT (1992) Relative rates of nucleotide substitution at the rbcL locus of the monocotyledonous plants. J Mol Evol 35:292–303
Gaut BS, Morton BR, McCaig BC, Clegg MT (1996) Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA 93:10274–10279
Gogoi B, Bhau BS (2018) DNA barcoding of the genus Nepenthes (Pitcher plant): a preliminary assessment towards its identification. BMC Plant Biol 18:153
Gunn BF (2004) The phylogeny of the Cocoeae (Arecaceae) with emphasis on Cocos nucifera. Ann Mo Bot Gard 91:505–522
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis: Department of Microbiology, North Carolina State University
Han YW, Duan D, Ma XF et al (2016) Efficient identification of the forest tree species in Aceraceae using DNA barcodes. Front Plant Sci 7:1707
Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B Biol Sci 270:313–321
Hebert PD, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the Neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA 101(41):14812–14817
Hollingsworth ML, Clark LA, Forrest LL, Richardson J, Pennington RT et al (2009) Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour 9:439–457
Hollingsworth PM, Graham SW, Little DP (2011) Choosing and using a plant DNA barcode. PLoS ONE 6:e19254
Jacob J, Mohanan N, Kariyappa K (2008) A new species of Calamus L. (Arecaceae) from Silent Valley, the Western Ghats, India. Rheedea 18:29–31
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–405
Jeanson ML, Labat JN, Little DP (2011) DNA barcoding: a new tool for palm taxonomists? Ann Bot 108:1445–1451
Joly S, Davies TJ, Archambault A et al (2014) Ecology in the age of DNA barcoding: the resource, the promise and the challenges ahead. Mol Ecol Resour 14:221–232
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kress WJ, Erickson DL (2007) A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2:e508
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102:8369–8374
Kuppu R, Manoharan S, Uthandakalaipandian R (2019) Phylogeographic study of freshwater fish Channa striata (Bloch) and Channa punctata (Bloch) construed using DNA barcodes between three rivers in southern Tamil Nadu region, India. Proc Zool Soc. https://doi.org/10.1007/s12595-019-00299-1
Kurian A (2018) Molecular systematics of rattans of south India. Doctoral thesis, Cochin University of Science and Technology, India, p 131
Lakshmana A, Renuka C (1990) New species of Calamus (Arecaceae) from India. JETBD 14:705–709
Lee HL, Yi DK, Kim JS, Kim KJ (2007) Development of plant DNA barcoding markers from the variable noncoding regions of chloroplast genome. The Second International Barcode of Life Conference, Taipei, Taiwan. pp. 18–20
Lewis CE, Doyle JJ (2001) Phylogenetic utility of the nuclear gene malate synthase in the palm family (Arecaceae). Mol Phylogenet Evol 19:409–420
Lewis CE, Doyle JJ (2002) Phylogenetic analysis of tribe Areceae (Arecaceae) using two low-copy nuclear genes. Plant Syst Evol 236:1–17
Lewis CE, Martinez N (2000) Identity of the Hyophorbe palms at the botanical garden of Cienfuegos, Cuba. Palms 44:93–97
Loo AHB, Dransfield J, Chase MW, Baker WJ (2006) Low-copy nuclear DNA, phylogeny and the evolution of dichogamy in the betel nut palms and their relatives (Arecinae; Arecaceae). Mol Phylogenet Evol 39:598–618
Maia VH, Mata CSd, Franco LO, Cardoso MA, Cardoso SRS et al (2012) DNA barcoding Bromeliaceae: achievements and pitfalls. PLoS ONE 7(1):e29877
Meier R, Shiyang K, Vaidya G, Ng PK (2006) DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol 55:715–728
Meyer CP, Paulay G (2005) DNA barcoding: error rates based on comprehensive sampling. PLoS Biol 3:e422
Moura C, Brambach F, Jair Hernandez Bado K et al (2019) Integrating DNA barcoding and traditional taxonomy for the identification of Dipterocarps in Remnant Lowland Forests of Sumatra. Plants (Basel, Switzerland) 8(11):461
Naeem A, Khan AA, Cheema HMN, Khan IA, Buerkert A (2014) DNA barcoding for species identification in the palmae family. Genet Mol Res 13:10341–10348
Newmaster SG, Fazekas A, Ragupathy S (2006) DNA barcoding in the land plants: evaluation of rbcL in a multigene tiered approach. Can J Bot 84:335–341
Norup MV (2006) A molecular systematic study of Heterospathe and Rhopaloblaste (Arecaceae, Areceae). Masters Thesis, University of Aarhus
Oxelman B, Bremer B (2000) Discovery of paralogous nuclear gene sequences coding for the second-largest subunit of RNA polymerase II (RPB2) and their phylogenetic utility in Gentianales of the asterids. Mol Biol Evol 17:1131–1145
Oxelman B, Yoshikawa N, Mcconaughy BL, Luo J, Denton AL, Hall BD (2004) RPB2 gene phylogeny in flowering plants, with particular emphasis on asterids. Mol Phylogenet Evol 32:462–479
Pang X, Song J, Zhu Y, Xu H, Huang L et al (2010) Applying plant DNA barcodes for Rosaceae species identification. Cladistics 27:165–170
Paracchini V, Petrillo M, Lievens A et al (2017) Novel nuclear barcode regions for the identification of flatfish species. Food Control 79:297–308
Percy DM, George WA, Quentin CC et al (2014) Understanding the spectacular failure of DNA barcoding in willows (Salix): does this result from a trans-specific selective sweep? Mol Ecol 19:4737–4756
Pfeil BE, Brubaker CL, Craven LA, Crisp MD (2004) Paralogy and orthology in the Malvaceae RPB2 gene family: investigation of gene duplication in hibiscus. Mol Biol Evol 21:1428–1437
Pillon Y, Johansen J, Sakishima T et al (2013) Potential use of low-copy nuclear genes in DNA barcoding: a comparison with plastid genes in two Hawaiian plant radiations. BMC Evol Biol 13:35
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808
Ran JH, Wang PP, Zhao HJ, Wang XQ (2010) A test of seven candidate barcode regions from the plastome in Picea (Pinaceae). J Integr Plant Biol 52:1109–1126
SPSS Inc. Released (2007) SPSS for Windows, Version 17.0. Chicago, SPSS Inc
Renuka C (1986) A new species of Calamus (Palmae) from India. Kew Bull 42:433–435
Renuka C (1999) Notes on the identity of Calamus delessertianus Becc. Rheedea 9:81–84
Renuka C (2001) Palms of India: status, threats and conservation strategies. In: Shaanker UR, Ganeshaiah KN, Bawa KS (eds) Forest genetic resources: status, threats and conservation strategies. Oxford & IBH Publishing Co. Pvt. Ltd., Calcutta, pp 197–209
Renuka C, Sasidharan N, Anto P (1997) A new species of Calamus (Arecaceae) from Silent Valley, Kerala, India. Rheedea 7:69–71
Renuka C, Bhat KV, Pandalai RC (2010) Rattans of India: taxonomy, biology and utilization. Kerala Forest Research Institute (KFRI), Thrissur, p 339
Roncal J, Francisco-Ortega J, Asmussen CB, Lewis CE (2005) Molecular phylogenetics of tribe Geonomeae (Arecaceae) using nuclear DNA sequences of phosphoribulokinase and RNA polymerase II. Syst Bot 30:275–283
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Roy S, Tyagi A, Shukla V et al (2010) Universal plant DNA barcode loci may not work in complex groups: a case study with Indian berberis species. PLoS ONE 5:e13674
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees”. Mol Biol Evol 4:406–425
Sang T (2002) Utility of low-copy nuclear gene sequences in plant phylogenetics. Crit Rev Biochem Mol Biol 37:121–147
Sass C, Little DP, Stevenson DW, Specht CD (2007) DNA barcoding in the Cycadales: testing the potential of proposed barcoding markers for species identification of Cycads. PLoS ONE 2(11):e1154
Scarcelli N, Barnaud A, Eiserhardt W, Treier UA, Seveno M, d’Anfray A, Vigouroux Y, Pintaud JC (2011) A set of 100 chloroplast primer pairs to study population genetics and phylogeny in monocotyledons. PLoS ONE 6:e19954
Senthilkumar U (2015) Phylogenetics and phylogeography of climbing palms of India (Tribe Calameae, Arecaceae). Doctoral thesis, Madras Christian College, India, p 205
Small R, Cronn R, Wendel J (2004) L.A.S Johnson Review No.2. Use of nuclear genes for phylogeny reconstruction in plants. Aust Syst Bot 17:145–170
Spooner DM (2009) DNA barcoding will frequently fail in complicated groups: an example in wild potatoes. Am J Bot 96:1177–1189
Sreekumar VB (2005) Systematics and phylogeny of the genus Calamus L. (Arecaceae) in the Western Ghats. Doctoral thesis, University of Calicut, India, p 140
Sreekumar VB, Henderson A (2014) Nomenclatural notes on Indian Calamus (Arecaceae). Phytotaxa 166:145–149
Stoeckle M (2003) Taxonomy, DNA, and the barcode of life. Bioscience 53(9):2–3
Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S (2012) Estimating divergence times in large molecular phylogenies. Proc Natl Acad Sci USA 109:19333–19338
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Techen N, Parveen I, Pan Z, Khan IA (2014) DNA barcoding of medicinal plant material for identification. Curr Opin Biotechnol 25:103–110
Thomas MM, Garwood NC, Baker WJ et al (2006) Molecular phylogeny of the palm genus Chamaedorea, based on the low-copy nuclear genes PRK and RPB2. Mol Phylogenet Evol 38:398–415
Uhl N, Dransfield J (1987) Genera Palmarum. Allen Press, Lawrence
Uhl NW, Dransfield J, Davis JI, Luckow MA, Hansen KS, Doyle JJ (1995) Phylogenetic relationships among palms: cladistic analyses of morphological and chloroplast DNA restriction site variation. In: Rudall PJ, Cribb PJ, Cutler DF, Humphries CJ (eds) Monocotyledons: systematics and evolution, vol 2. Royal Botanic Gardens, Kew, pp 623–661
Umapathy S, Duvuru N, Munivenkatappa S, Uma Shaanker R, Gudasalamani R (2015) Species delimitation in congenerics of genus Daemonorops from India using DNA barcodes. Commun Plant Sci 5(1–2):1–8
Valentini A, Miquel C, Nawaz MA et al (2009) New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: the trnL approach. Mol Ecol Resour 9:51–60
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Wilson MA, Gaut B, Clegg MT (1990) Chloroplast DNA evolves slowly in the palm family (Arecaceae). Mol Biol Evol 7:303–314
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast and nuclear DNAs. Proc Natl Acad Sci USA 84:9054–9058
Yang HQ, Dong YR, Gu ZJ, Liang N, Yang JB (2012) A preliminary assesment of matK, rbcL and trnH-psbA as DNA barcodes for Calamus (Arecaceae) species in China with a note on ITS. Ann Bot Fenn 49:319–330
Yu J, Xue JH, Zhou SL (2011) New universal matK primers for DNA barcoding angiosperms. J Syst Evol 49:176–181
Zona S, Ortega JF, Jestrow B, Baker WJ, Lewis CE (2011) Molecular phylogenetics of the palm subtribe Ptychospermatinae (Arecaceae). Am J Bot 98:1716–1726
Acknowledgements
We thank Kerala Forest Department, Govt. of Kerala and Karnataka Forest Department, Govt. of Karnataka for their permission to collect samples. The financial support received from Kerala State Council for Science Technology and Environment (KSCSTE) is also acknowledged.
Funding
The financial support for this study was received from Kerala State Council for Science, Technology and Environment (KSCSTE) (Grant No. KFRI RP 617/2011).
Author information
Authors and Affiliations
Contributions
AK: contributed towards conducting wet lab experiments, sample collections from the field, writing of the paper and analysing the data, SAD: involved in writing of the paper and designing the project, SVB: sample collections from the field site, identification and writing of the paper, MEM: involved in coordination of project and writing of the paper
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig.1.
PCR amplified products of the analysed DNA barcode loci (TIFF 202 kb)
Supplementary Fig.2.
Multiple Sequence Alignment showing species specific differences in the Calamus species using RPB2 (TIFF 2057 kb)
Supplementary Fig.3.
Multiple Sequence Alignment showing 300bp deletion in the C. hookerianus and C. pseudotenuis using psbA-trnH. (TIFF 580 kb)
Supplementary Fig.4.
Multiple Sequence Alignment showing transversions in the Calamus gamblei species complex (TIFF 1553 kb)
Supplementary Fig.5.
Neighbor-joining tree (NJ) of the genus Calamus using RPB2 based on p-distance using MEGA v.6.0 with bootstrap percentage value shown below (TIFF 53 kb)
Supplementary Fig.6.
Phylogenetic tree with posterior probability using RPB2 of the genus Calamus (TIFF 553 kb)
Supplementary Fig.7.
Phylogenetic tree with posterior probability using RPB2 of the family Arecaceae (TIFF 114 kb)
Supplementary Table S1.
List of samples collected for DNA Barcoding, their location and GenBank Accession numbers (DOC 118 kb)
Supplementary Table S2.
List of sequences downloaded from GenBank and their Accession numbers. (DOC 29 kb)
Supplementary File 1.
Multiple Sequence Alignment of RPB2 (MAS 50 kb)
Rights and permissions
About this article
Cite this article
Kurian, A., Dev, S.A., Sreekumar, V.B. et al. The low copy nuclear region, RPB2 as a novel DNA barcode region for species identification in the rattan genus Calamus (Arecaceae). Physiol Mol Biol Plants 26, 1875–1887 (2020). https://doi.org/10.1007/s12298-020-00864-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-020-00864-5