Abstract
Key message
SSRs related to parent selection of D. kaki were validated by transcriptome sequencing, and genetic diversity of these markers among 76 persimmon germplasm resources was additionally investigated.
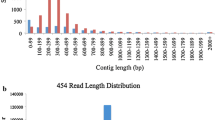
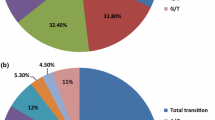
The selection of hybrid parents is critical in cross-breeding plants for germplasm innovation and cultivating improved varieties. This study used a published RNA-Seq database of a persimmon cultivar ‘Zenjimaru’ to develop simple sequence repeat (SSR) markers to assist efficiently selection of appropriate persimmon cultivars for cross-breeding. A total of 154,741 unigenes with 41,999 SSR loci at a frequency of one per 2.30 kb were identified. Of the different SSR motif types, mononucleotide repeats (47.63%) were the most abundant, followed by dinucleotide repeats (38.23%). The main repeat motifs were A/T (42.26%) and AG/CT (24.72%). A total of 17,111 SSR primer pairs were designed, from which 100 selected loci were synthesized and preliminary screened with capillary electrophoresis by testing PCR products from 15 persimmon germplasm resources. The effectiveness of 13 primer pairs that produced products with distinct peaks at the predicted sizes was subsequently determined using 76 persimmon germplasm resources. A total of 111 alleles were identified by using 13 SSR markers, ranging from 2 to 17 with an average of 8.54 bands per locus. Additionally, different hybridized combinations could be selected according to a dendrogram based on these newly developed 13 SSR markers. And two androecious persimmons (“Yunjia-9” and “Luotian Yeshi-1”) were genetically closely related to Chinese pollination-constant non-astringent (C-PCNA) persimmons, which was significant for the cultivation of PCNA offspring. The identified SSR markers will not only promote persimmon cross-breeding programs by increasing the efficiency of parent selection but also assist in genetic resource identification and conservation.




Similar content being viewed by others
References
Akagi T, Katayama-Ikegamib A, Yonemoria K (2011) Proanthocyanidin biosynthesis of persimmon (Diospyros kaki Thunb.) fruit. Sci Hortic 130(2):373–380
Akagi T, Henry IM, Tao R, Comai L (2014) A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science 346(6209):646–650
Akagi T, Henry IM, Kawai T, Comai L, Tao R (2016) Epigenetic regulation of the sex determination gene MeGI in polyploid persimmon. Plant Cell 28(1):2905–2915
Bematzky R, Mulcahy DL, Tuskan GA (1992) Marker-aided selection in a backcross breeding program for resistance to chestnut blight in the American chestnut. Can J For Res 22(7):1031–1035
Bhargava A, Fuentes FF (2010) Mutational dynamics of microsatellites. Mol Biochnol 44(3):250–266
Chen J, Li R, Xia Y, Bai G, Guo P, Wang Z, Zhang H, Siddique KHM (2017) Development of EST-SSR markers in flowering Chinese cabbage (Brassica campestris L. ssp. chinensis var. utilisTsen et Lee) based on de novo transcriptomic assemblies. PLoS ONE 12(9):e0184736
Deng KP, Deng RJ, Fan JX, Chen EF (2018) Transcriptome analysis and development of simple sequence repeat (SSR) markers in Zingiber striolatum Diels. Physiol Mol Biol Plants 24(1):125–134
Ding Y, Xue L, Guo RX, Luo GJ, Song YT, Lei JJ (2019) De novo assembled transcriptome analysis and identification of genic SSR markers in red-flowered strawberry. Biochem Genet 57(5):607–622
Dong L, Sun YH, Zhao KQ, Zhang J, Zhang YW, Li XY, Xun SH, Zhang JT, Wang SM, Li Y (2019) Development and application of EST-SSR markers for DNA fingerprinting and genetic diversity analysis of the main cultivars of black locust (Robinia pseudoacacia L.) in China. Forests 10(8):644
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Du F, Wu Y, Zhang L, Li XW, Zhao XY, Wang WH, Gao ZS, Xia YP (2015) De novo assembled transcriptome analysis and SSR marker development of a mixture of six tissues from Lilium Oriental hybrid ‘Sorbonne’. Plant Mol Biol Rep 33(2):281–293
Gao Z, Wu J, Liu Z, Wang L, Ren H, Shu Q (2013) Rapid microsatellite development for tree peony and its implications. BMC Genomics 14(1):886
George AP, Mowat AD, Collins RJ, Morley-Bunker M (1997) The pattern and control of reproductive development in nonastringent persimmon (Diospyros kaki L.): a review. Sci Hortic 70(2–3):93–122
Guan CF, Chen WX, Mo RL, Du XY, Zhang QL, Luo ZR (2016) Isolation and characterization of DkPK genes associated with natural deastringency in C-PCNA persimmon. Front Plant Sci 7:1–11
Guo DL, Luo ZR (2006) Genetic relationships of some PCNA persimmons (Diospyros kaki Thunb.) from China and Japan revealed by SRAP analysis. Genet Res Crop Evol 53:1597–1603
Guo DL, Luo ZR (2008) Microsatellite isolation and characterization in Japanese persimmon (Diospyros kaki). Biochem Genet 46:323–328
Guo Q, Wang JX, Su LZ, Lv W, Sun YH, Li Y (2017) Development and evaluation of a novel set of EST-SSR markers based on transcriptome sequences of black locust (Robinia pseudoacacia L.). Genes 8(7):177
Ikegami A, Eguchi S, Sato A, Yamada M, Kitajima A, Mitani N (2006) Segregations of astringent progenies in the F1 populations derived from crosses between a Chinese pollination constant non-astringent (PCNA) ‘Luo Tian Tian Shi’, and Japanese PCNA and pollination-constant, astringent (PCA) cultivars. Hort Sci 41(3):561–563
Ikegami A, Eguchi S, Akagi T, Sato A, Yamada M, Kanzaki S (2011) Development of molecular markers linked to the allele associated with the non-astringent trait of Chinese persimmon (Diospyros kaki Thunb.). J Japan Soc Hortic Sci 80(2):150–155
Ismail NA, Rafii MY, Mahmud TMM, Hanafi MM, Miah G (2016) Molecular markers: a potential resource for ginger genetic diversity studies. Mol Biol Rep 43(12):1347–1358
Kajiura M (1946) Persimmon cultivars and their improvement. Breed Hortic 1:175–182
Kantety RV, La RM, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48(5–6):501–510
Kumpatla SP, Mukhopadhyay S (2005) Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species. Genome 48(6):985–998
Li SZ, Sun P, Du GG, Wang LY, Li HW, Fu JM, Suo YJ, Han WJ, Diao SF, Mai YN, Li FD (2019) Transcriptome sequencing and comparative analysis between male and female floral buds of the persimmon (Diospyros kaki Thunb.). Sci Hortic 246:987–997
Liang X, Chen X, Hong Y, Liu H, Zhou G, Li S, Guo B (2009) Utility of EST-derived SSR in cultivated peanut (Arachis hypogaea L.) and Arachis wild species. BMC Plant Biol 9(1):1–9
Liang YQ, Han WJ, Sun P (2015) Genetic diversity among germplasms of Diospyros kaki based on SSR markers. Sci Hortic 186:180–189
Liu F, Wang YS, Tian XL, Mao ZC, Zou XX, Xie BY (2012) SSR mining in pepper (Capsicum annuum L.) transcriptome and the polymorphism analysis. Acta Hortic Sin 39(1):168–174
Luo ZR, Wang RZ (2008) Persimmon in China: domestication and traditional utilizations of genetic resources. Adv Hortic Sci 22:239–243
Luo ZR, Li FF, Cai LH (1999) Molecular systematics of China native nonastringent persimmon based on random amplified polymorphic DNA. Acta Hort Sinica 26(5):297–301
Luo C, Zhang F, Zhang QL, Guo DY, Luo ZR (2013) Characterization and comparison of EST-SSR and TRAP markers for genetic analysis of the Japanese persimmon Diospyros kaki. Genet Mol Res 12(3):2841–2851
Nakagawa T, Nakatsuka A, Yano K, Yasugahira S, Nakamura R, Sun N, Itai A, Suzuki T, Itamura H (2008) Expressed sequence tags from persimmon at different developmental stages. Plant Cell Rep 27:931–938
Sablok G, Luo C, Lee WS, Rahman F, Tatarinova TV, Harikrishna JA, Luo ZR (2011) Bioinformatic analysis of fruit-specific expressed sequence tag libraries of Diospyros kaki Thunb.: view at the transcriptome at different developmental stages. 3Biotech 1:35–45
Soriano JM, Pecchioli S, Romero C, Vilanova S, Llácer G, Badenes ML (2006) Development of microsatellite markers in polyploid persimmon (Diospyros kaki Lf) from an enriched genomic library. Mol Ecol Notes 6:368–370
Spongberg SA (1979) Notes on persimmons, kakis, date plums and chapotes. Arnoldia 39(5):290–309
Sugiura A, Tomana T (1983) Relationships of ethanol production by seeds of different types of Japanese persimmons and their tannin content. Hort Sci 18:319–321
Ujino T, Kawahara T, Tsumura Y, Nagamitsu T, Yoshimaru H, Ratnam W (1999) Development and polymorphism of simple sequence repeat DNA markers for Shorea curtisii, and other dipterocarpaceae species. Heredity 81(4):422–428
Uzunova MI, Ecke W (1999) Abundance, polymorphism and genetic mapping of microsatellites in oilseed rape (Brassica napus L.). Plant Breeding 118(4):323–326
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23(1):48–55
Wang CB, Guo WZ, Cai CP, Zhang TZ (2006) Characterization, development and exploitation of EST derived microsatellites in Gossypium raimondii Ulbrich. Chin Sci Bull 51(5):557–561
Wang J, Lu C, Yuan C, Cui B, Qiu Q, Sun P, Hu R, Wu D, Sun Y, Li Y (2015) Characterization of ESTs from black locust for gene discovery and marker development. Genet Mol Res 14:12684–12691
Wang LY, Li HW, Sun P, Fu JM, Suo YJ, Zhang JJ, Han WJ, Diao SF, Li FD, Mai YN (2018) Genetic diversity among wild androecious germplasms of Diospyros kaki in China based on SSR markers. Sci Hortic 242:1–9
Wang L, Gong XQ, Jin L, Li HY, Lu JQ, Duan J, Ma LY (2019) Development and validation of EST-SSR makers of Magnolia wufengensis using de novo transcriptome sequencing. Trees 33(4):1213–1223
Xu LQ, Zhang QL, Luo ZR (2013) Pollen-related characteristics of Diospyros Linn. (Ebenaceae) androecious germplasms newly found in China. Acta Hortic 996:199–206
Yamada M, Sato A, Yakushiji H, Yashinaga K, Yamane H, Endo M (1993) Characteristic of ‘Luo Tian Tian Shi’, a non-astringent cultivar of oriental persimmon (Diospyros kaki Thunb.) of Chinese origin in relation to non-astringent cultivars of Japanese origin. Bull Fruit Tree Res Stn 25:19–32
Yang HS, Li XP, Liu DJ, Chen XB, Li FH, Qi XL, Luo ZW, Wang CB (2016) Genetic diversity and population structure of the endangered medicinal plant Phellodendron amurense in China revealed by SSR markers. Biochem Syst Ecol 66:286–292
Yonemori K, Sugiura A, Tanaka K, Kameda K (1993) Floral ontogeny and sex determination in monoecious-type persimmons. J Am Soc Hortic Sci 118(2):293–297
Zhang QL, Guo DL, Luo ZR (2009) Identification and taxonomic status of Chinese Diospyros spp. (Ebenaceae) androecious germplasms. Acta Hortic 833:91–96
Zhang QT, Li XY, Yang YM, Fan ST, Ai J (2016a) Analysis on SSR information in transcriptome and development of molecular markers in Lonicera caerulea. Acta Hortic Sin 43(3):557–563
Zhang N, Mo RL, Zhang QL, Luo ZR (2016b) Application potential of Chinese androecious genotype, ‘Male 8’ as the parent in the genetic improvement of PCNA persimmon. Acta Horti Sin 43(11):2133–2140 (In Chinese)
Zhang PX, Yang SC, Liu YF, Zhang QL, Xu LQ, Luo ZR (2016c) Validation of a male-linked gene locus (OGI) for sex identification in persimmon (Diospyros kaki Thunb.) and its application in F1 progeny. Plant Breeding 135(6):721–727
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (2019YFD1001200) and the Fundamental Research Funds for the Central Non-profit Research Institution of CAF (CAFYBB2017ZA005, CAFYBB2019GC001-19, and CAFYBB2017ZA004-3).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by M. Fladung.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, L., Li, H., Suo, Y. et al. Development of EST-SSR markers and their application in the genetic diversity of persimmon (Diospyros kaki Thunb.). Trees 35, 121–133 (2021). https://doi.org/10.1007/s00468-020-02024-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-020-02024-4