Abstract
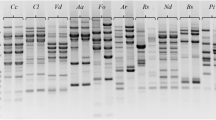
A total of 76 Corynespora cassiicola isolates collected from 16 rubber clones in several geographic regions of Vietnam, were studied for genetic diversity by ribosomal DNA internal transcribed spacer (rDNA-ITS) sequences and sequence-related amplified polymorphism (SRAP) techniques. Results of rDNA-ITS sequence analysis indicated 2 nucleotide polymorphisms at the 135th (cytosine/thymine) and 474th (guanine/adenine) base pair positions of rDNA-ITS region, which differentiated the 76 studied isolates into 3 clusters. All isolates could be uniquely distinguished by 208 polymorphism bands which generated 93.3% polymorphic ratio with SRAP markers. A dendrogram produced from UPGMA analysis based on Nei and Li’s coefficient divided the 76 C. cassiicola isolates into 2 main clusters. Cluster one included 54 fungal isolates of which 51 and 3 were observed in subgroup 1A and 1B, respectively. There were 22 C. cassiicola isolates belonging to cluster 2 with subgroups 2A and 2B consisted of 20 and 2 fungal isolates, respectively. Bootstrap values for groups one and two were 61% and 100%, respectively and the grouping was reliable, with similarity coefficient between the two main groups at 67%. rDNA-ITS sequence analysis revealed a new genetic group, signifying at least three C. cassiicola genetic groups that infect rubber trees in Vietnam. SRAP markers divided the studied isolates into two distinct groups, which correlated with geographical location rather than host source. To the best of our knowledge, this is the first demonstration using SRAP markers on the systematics of C. cassiicola isolates which cause the Corynespora leaf fall disease on various rubber (Hevea brasiliensis) clones in Vietnam.







Similar content being viewed by others
References
Farr DF, Rossman AY (2019) Fungal Databases, Systematic Mycology and Microbiology Laboratory, ARS, USDA. https://nt.ars-grin.gov/fungaldatabases/. Retrieved March 12, 2019
Déon M, Fumanal B, Gimenez S, Bieysse D, Olivera RR, Shuib SS, Breton F, Elumalai S, Vida JB, Seguin M, Leroy T, Roeckel-Drevet P, Pujade-Renaud V (2014) Diversity of the cassiicolin gene in Corynespora cassiicola and relation with the pathogenicity in Hevea brasiliensis. Fungal Biology 118(1):32–47
Damono TW, Darussamin A, Pawirosoemardjo S (1996) Variation among isolates of Corynespora cassiicola associated with Hevea brasiliensis in Indonesia. Proceeding Workshop on Corynespora Leaf Fall Disease of Hevea Rubber. Medan, Indonesia. Indonesian Rubber Research Institute, pp 79–91
Saha T, Kumar A, Sreena AS, Joseph A, Jacob CK, Kothandaraman R, Nazeer MA (2000) Genetic variability of Corynespora cassiicola infecting Hevea brasiliensis isolated from the traditional rubber growing areas in India. Indian J Nat Rubb Res 13:1–10
Silva WPK, Karunanayake EH, Ravi LCW, Priyanka UMS (2003) Genetic variation in Corynespora cassiicola: a possible relation between host origin and virulence. Mycol Res 107(5):567–571
Nghia NA, Kadir J, Sunderasan E, Abdullah MP, Malik A, Napis S (2008) Morphorlogical and Inter Simple Sequence Repeat (ISSR) Markers Analyses of Corynespora cassiicola Isolates from Rubber Plantations in Malaysia. Mycopathologia 166:189–201
Qi YX, Xie YX, Zhang X, Pu J, Zhang HQ, Huang S, Zhang H (2009) Molecular and pathogenic variation identified among isolates of Corynespora cassiicola. Mol Biotechnol 41(2):145–151
Hieu ND, Nghia NA, Chi VTQ, Dung PT (2014) Genetic diversity and pathogenicity of Corynespora cassiicola isolates from rubber tree and other hosts in Vietnam. J Rubb Res 73(3):187–203
Atan T, Hamid NH (2003) Differentiating races of Corynespora cassiicola using RAPD and internal transcribed spacer markers. J Rubb Res 6(1):58–64
Fernando THPS, Jayasinghe CK, Wijesundera RLC, Siriwardana D (2009) Variability of Hevea isolate of Corynespora cassiicola from Sri Lanka. J Plant Dis Protect 116(3):115–117
Atan S, Derapi S, Ismail L, Shukor NAA (2011) Screening susceptibility of Hevea progenies from PB 5/51 X IAN 873 to two races of Corynespora cassiicola. J Rubb Res 14(2):110–122
Silva WPK, Deveral BJ, Lyon BR (1998) Molecular, physiological and pathological characterization of Corynespora leaf spot fungi from rubber plantations in Sri Lanka. Plant Pathol 3:267–277
Dixon LJ, Schulub RL, Pernezny K, Datnoff LE (2009) Host specialization and phylogenetic diversity of Corynespora cassiicola. Phytopathology 99:1015–1027
Romruensukharom P, Tragoonrung S, Vanavichit A, Toojinda T (2005) Genetic variability of Corynespora cassiicola population in Thailand. J Rubb Res 8:38–49
Shimomoto Y, Sato T, Hojo H, Morita Y, Takeuchi S, Mizumoto H, Kiba A, Hikichi Y (2010) Pathogenic and genetic variation among isolates of Corynespora cassiicola in Japan. Plant Pathol 60:253–260
Yao FJ, Lu LX, Wang P, Fang M, Zhang YM, Chen Y, Zhang WT, Kong XH, Lu J, Honda Y (2018) Development of a molecular marker for fruiting body pattern in Auricularia auricula-judae. Mycobiology 46:72–78
Liu J, Wang ZR, Li C, Bian YB, Xiao Y (2015) Evaluating genetic diversity and constructing core collections of Chinese Lentinula edodes cultivars using ISSR and SRAP markers. J Basic Microbiol 55:749–760
Zhang QS, Xu BL, Liu LD, Yuan QQ, Dong HX, Cheng XH, Lin DL (2012) Analysis of genetic diversity among Chinese Pleurotu scitrinopileatus singer cultivars using two molecular marker systems (ISSRs and SRAPs) and morphological traits. World J Microbiol Biotechnol 28:2237–2248
Hasan HA, Almomany AM, Hasan S, Al-Abdallat AM (2018) Assessment of genetic diversity among Pleurotus spp. isolates from Jordan., Journal of Fungi (Basel) 4, 52.
Yin Y, Liu Y, Li H, Zhao S, Wang S, Liu Y, Wu D, Xu F (2014) Genetic diversity of Pleurotus pulmonarius revealed by RAPD, ISSR, and SRAP fingerprinting. Curr Microbiol 68:397–403
Ma DL, Yang GT, Mu LQ, Song YT (2010) Application of SRAP in the genetic diversity of Tricholoma matsutake in Northeastern China. African J Biotechnol 9:6244–6250
Li XY, Li J, Zhao ZJ, Yang F, Fu QW, Liu HS, Wang DD, Yang YC, Wang RY (2014) Sequence-related amplified polymorphism (SRAP) for studying genetic diversity and population structure of plants and other living organisms: a protocol. J Anim Plant Sci 24(5):1478–1486
Ferriol M, PicoB NF (2003) Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP markers. Theor Appl Genetics 107:271–282
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Hall TA (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Tamara K, Steche G, Peterson D, Filipski D, Sudhir Kumar S (2013) MEGA6: Molecular evolutionary genetic analysis version 6.0. Mol Biol Evol 30:2725–2729
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), A new marker system based on a simple PCR reaction: Its application to mapping and gene tagging in Brassica. Theor Appl Genet 103:455–461
Pavlíček A, Hrdá Š, Flegr J (1999) FreeTree - Freeware program for construction of phylogenetic trees on the basis of distance data and for Bootstrap/Jackknife analysis of the trees robustness. Application in the RAPD analysis of Genus Frenkelia. Folia Biol 45:97–99
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Gil-Lamaignere C, Roilides E, Hacker J, Muller FMC (2003) Molecular typing for fungi—a critical review of the possibilities and limitation of currently and future methods. Clin Microbiol Infect 9:172–185
Nghia NA, Kadir J, Sunderasan E, Puad Abdullah M, Napis S (2010) Intraspecific variability of Corynespora cassiicola inferred from single nucleotide polymorphisms in ITS Region of Ribosomal DNA. J Rubb Res 13(4):257–264
Qi YX, Zhang X, Pu JJ, Liu XM, Lu Y, Zhang H, Zhang HQ, Lu YC, Xie YX (2011) Morphological and molecular analysis of genetic variability within isolates of Corynespora cassiicola from different hosts. Eur J Plant Pathol 130:83–95
Oktavia F, Kuswanhadi W, Dinarti D, Sudarsono S (2017) Pathogenicity and rDNA-ITS sequence analysis of the Corynespora cassiicola isolates from rubber plantations in Indonesia. Emi J Food Agric 29(11):872–883
Xu L, Lu Y, You Q, Liu X, Grisham MP, Pan Y, Que Y (2014) Biogeographical variation and population genetic structure of Sporisorium scitamineum in mainland in China insights from ISSR and SP-SRAP markers. Sci World J. https://doi.org/10.1155/2014/296020
Que Y, Xu L, Lin J, Chen R, Grisham MP (2012) Molecular variation of Sporisorium scitamineum in mainland China revealed by RAPD and SRAP markers. Plant Dis 96(10):1519–1525
Chen BY, Hu Q, Dixelius C, Li GQ, Wu XM (2010) Genetic diversity in Sclerotinia sclerotiorum assessed with SRAP markers. Biodivers Sci 18(5):509–515
Soren KR, Gangwar P, Khatterwani P, Chaudhary RG, Datta S (2016) Genetic diversity assessment of Fusarium oxysporum f.sp.ciceris isolates of Indian chickpea fields as revealed by the SRAP marker system. J Environ Biol 37(6):1291–97
Tian YH, Hou YY, Peng CY, Wang YY, He BL, Gao KX (2017) Genetic diversity and phylogenetic analysis of Fusarium oxysporium strains isolated from the Cucurbitaceae hosts revealed by SRAPs. Ying Yong Sheng Tai Xue Bao 28(3):947–956
Acknowledgements
We are grateful to the Rubber Research Institute of Vietnam and the Research Institute for Biotechnology and Environment, Nong Lam University, Vietnam for supporting this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors state no conflict of interest related to this publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hieu, N.D., Nghia, N.A., Uyen, N.T.K. et al. Genetic diversity analysis of Corynespora cassiicola isolates on the rubber tree (Hevea brasiliensis) in Vietnam using ribosomal DNA internal transcribed spacer (rDNA-ITS) sequences and sequence-related amplified polymorphism (SRAP). J Rubber Res 23, 173–185 (2020). https://doi.org/10.1007/s42464-020-00047-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42464-020-00047-7