Abstract
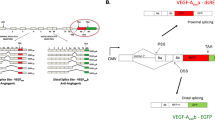
Vascular endothelial growth factor A (VEGFA) stimulates angiogenesis in human endothelial cells, and increasing its expression is a potential treatment for heart failure. Here, we report the design of a small molecule (TGP-377) that specifically and potently enhances VEGFA expression by the targeting of a non-coding microRNA that regulates its expression. A selection-based screen, named two-dimensional combinatorial screening, revealed preferences in small-molecule chemotypes that bind RNA and preferences in the RNA motifs that bind small molecules. The screening program increased the dataset of known RNA motif–small molecule binding partners by 20-fold. Analysis of this dataset against the RNA-mediated pathways that regulate VEGFA defined that the microRNA-377 precursor, which represses Vegfa messenger RNA translation, is druggable in a selective manner. We designed TGP-377 to potently and specifically upregulate VEGFA in human umbilical vein endothelial cells. These studies illustrate the power of two-dimensional combinatorial screening to define molecular recognition events between ‘undruggable’ biomolecules and small molecules, and the ability of sequence-based design to deliver efficacious structure-specific compounds.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting this manuscript are contained within the main text, source data and Supplementary figures. Specific data are freely available upon reasonable request from the corresponding author. The Inforna database9 can be accessed via the following URL https://disney.florida.scripps.edu/software/. Users wishing to obtain access must complete a software license agreement with TSRI, upon which login credentials will be provided after approval.
Code availability
No unique code was used in the described data analyses.
References
Angelbello, A. J. et al. Using genome sequence to enable the design of medicines and chemical probes. Chem. Rev. 118, 1599–1663 (2018).
Disney, M. D. & Angelbello, A. J. Rational design of small molecules targeting oncogenic noncoding RNAs from sequence. Acc. Chem. Res. 49, 2698–2704 (2016).
Stein, C. A. & Castanotto, D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 25, 1069–1075 (2017).
Stojic, L. et al. Specificity of RNAi, LNA and CRISPRi as loss-of-function methods in transcriptional analysis. Nucleic Acids Res. 46, 5950–5966 (2018).
Watters, K. E., Abbott, T. R. & Lucks, J. B. Simultaneous characterization of cellular RNA structure and function with in-cell SHAPE-Seq. Nucleic Acids Res. 44, e12 (2016).
Strobel, E. J., Yu, A. M. & Lucks, J. B. High-throughput determination of RNA structures. Nat. Rev. Genet. 19, 615–634 (2018).
Velagapudi, S. P., Gallo, S. M. & Disney, M. D. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat. Chem. Biol. 10, 291–297 (2014).
Tran, T. & Disney, M. D. Identifying the preferred RNA motifs and chemotypes that interact by probing millions of combinations. Nat. Commun. 3, 1125 (2012).
Disney, M. D. et al. Inforna 2.0: a platform for the sequence-based design of small molecules targeting structured RNAs. ACS Chem. Biol. 11, 1720–1728 (2016).
Ferrara, N., Gerber, H.-P. & LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 9, 669–676 (2003).
Sun, N. et al. Modified VEGF-A mRNA induces sustained multifaceted microvascular response and accelerates diabetic wound healing. Sci. Rep. 8, 17509–17519 (2018).
Ylä-Herttuala, S., Rissanen, T. T., Vajanto, I. & Hartikainen, J. Vascular endothelial growth factors: biology and current status of clinical applications in cardiovascular medicine. J. Am. Coll. Cardiol. 49, 1015–1026 (2007).
Taimeh, Z., Loughran, J., Birks, E. J. & Bolli, R. Vascular endothelial growth factor in heart failure. Nat. Rev. Cardiol. 10, 519–530 (2013).
Velagapudi, S. P. et al. Approved anti-cancer drugs target oncogenic non-coding RNAs. Cell Chem. Biol. 25, 1086–1094 (2018).
Velagapudi, S. P. et al. Defining RNA–small molecule affinity landscapes enables design of a small molecule inhibitor of an oncogenic noncoding RNA. ACS Cent. Sci. 3, 205–216 (2017).
Morgan, B. S., Forte, J. E., Culver, R. N., Zhang, Y. & Hargrove, A. E. Discovery of key physicochemical, structural, and spatial properties of RNA-targeted bioactive ligands. Angew. Chem. Int. Ed. 56, 13498–13502 (2017).
Mehta, A. et al. SMMRNA: a database of small molecule modulators of RNA. Nucleic Acids Res. 42, D132–D141 (2014).
Cordella, L. P., Foggia, P., Sansone, C. & Vento, M. A (sub)graph isomorphism algorithm for matching large graphs. IEEE Trans. Pattern Anal. Mac. Intell. 26, 1367–1372 (2004).
Ehrlich, H.-C. & Rarey, M. Systematic benchmark of substructure search in molecular graphs - from Ullmann to VF2. J. Cheminform. 4, 13 (2012).
Haniff, H. S., Graves, A. & Disney, M. D. Selective small molecule recognition of RNA base pairs. ACS Comb. Sci. 20, 482–491 (2018).
Schneider, T. D. & Stephens, R. M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990).
Stelzer, A. C., Kratz, J. D., Zhang, Q. & Al-Hashimi, H. M. RNA dynamics by design: biasing ensembles towards the ligand-bound state. Angew. Chem. Int. Ed. 49, 5731–5733 (2010).
Stelzer, A. C. et al. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat. Chem. Biol. 7, 553–559 (2011).
Chen, J. L., VanEtten, D. M., Fountain, M. A., Yildirim, I. & Disney, M. D. Structure and dynamics of RNA repeat expansions that cause Huntington’s disease and myotonic dystrophy type 1. Biochemistry 56, 3463–3474 (2017).
Childs-Disney, J. L. et al. A massively parallel selection of small molecule-RNA motif binding partners informs design of an antiviral from sequence. Chem 4, 2384–2404 (2018).
Costales, M. G. et al. Small molecule inhibition of microRNA-210 reprograms an oncogenic hypoxic circuit. J. Am. Chem. Soc. 139, 3446–3455 (2017).
Velagapudi, S. P. et al. Design of a small molecule against an oncogenic noncoding RNA. Proc. Natl Acad. Sci. USA 113, 5898–5903 (2016).
Wen, Z. et al. MicroRNA-377 regulates mesenchymal stem cell-induced angiogenesis in ischemic hearts by targeting VEGF. PLoS ONE 9, e104666 (2014).
el Azzouzi, H. et al. The hypoxia-inducible microRNA cluster miR-199a∼214 targets myocardial PPARδ and impairs mitochondrial fatty acid oxidation. Cell Metab. 18, 341–354 (2013).
Duan, Q. et al. MicroRNA-214 is upregulated in heart failure patients and suppresses XBP1-mediated endothelial cells angiogenesis. J. Cell. Physiol. 230, 1964–1973 (2015).
Wang, R., Dong, L.-D., Meng, X.-B., Shi, Q. & Sun, W.-Y. Unique microRNA signatures associated with early coronary atherosclerotic plaques. Biochem. Biophys. Res. Commun. 464, 574–579 (2015).
Yan, X. C. et al. MiR-342-5p is a notch downstream molecule and regulates multiple angiogenic pathways including Notch, vascular endothelial growth factor and transforming growth factor β signaling. J. Am. Heart Assoc. 5, e003042 (2016).
DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011).
Renton, A. E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011).
Brčić, J. & Plavec, J. NMR structure of a G-quadruplex formed by four d(G4C2) repeats: insights into structural polymorphism. Nucleic Acids Res. 46, 11605–11617 (2018).
Su, Z. et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 84, 1043–1050 (2014).
Wang, Z. F. et al. The hairpin form of r(G4C2)exp in c9ALS/FTD is repeat-associated non-ATG translated and a target for bioactive small molecules. Cell Chem. Biol. 26, 179–190 (2019).
Balendra, R. & Isaacs, A. M. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat. Rev. Neurol. 14, 544–558 (2018).
Ferrara, N., Hillan, K. J. & Novotny, W. Bevacizumab (Avastin), a humanized anti-VEGF monoclonal antibody for cancer therapy. Biochem. Biophys. Res. Commun. 333, 328–335 (2005).
Ivy, S. P., Wick, J. Y. & Kaufman, B. M. An overview of small-molecule inhibitors of VEGFR signaling. Nat. Rev. Clin. Oncol. 6, 569–579 (2009).
Ahmed, S. I., Thomas, A. L. & Steward, W. P. Vascular endothelial growth factor (VEGF) inhibition by small molecules. J. Chemother. 16, 59–63 (2004).
Arcondéguy, T., Lacazette, E., Millevoi, S., Prats, H. & Touriol, C. VEGF-A mRNA processing, stability and translation: a paradigm for intricate regulation of gene expression at the post-transcriptional level. Nucleic Acids Res. 41, 7997–8010 (2013).
Koransky, M. L., Robbins, R. C. & Blau, H. M. VEGF gene delivery for treatment of ischemic cardiovascular disease. Trends Cardiovasc. Med. 12, 108–114 (2002).
Liu, B. et al. Analysis of secondary structural elements in human microRNA hairpin precursors. BMC Bioinf. 17, 112–120 (2016).
Agarwal, V., Bell, G. W., Nam, J. W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005 (2015).
Holmes, K., Roberts, O. L., Thomas, A. M. & Cross, M. J. Vascular endothelial growth factor receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell. Signal. 19, 2003–2012 (2007).
Mao, Z., Liu, C., Lin, X., Sun, B. & Su, C. PPP2R5A: a multirole protein phosphatase subunit in regulating cancer development. Cancer Lett. 414, 222–229 (2018).
Angelbello, A. J. et al. Precise small-molecule cleavage of an r(CUG) repeat expansion in a myotonic dystrophy mouse model. Proc. Natl Acad. Sci. USA 116, 7799–7804 (2019).
Gan, L. M. et al. Intradermal delivery of modified mRNA encoding VEGF-A in patients with type 2 diabetes. Nat. Commun. 10, 871–879 (2019).
Zangi, L. et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 31, 898–907 (2013).
Hassan, M., Brown, R. D., Varma-O’Brien, S. & Rogers, D. Cheminformatics analysis and learning in a data pipelining environment. Mol. Divers. 10, 283–299 (2006).
Costales, M. G., Matsumoto, Y., Velagapudi, S. P. & Disney, M. D. Small molecule targeted recruitment of a nuclease to RNA. J. Am. Chem. Soc. 140, 6741–6744 (2018).
Acknowledgements
This work was funded by the National Institutes of Health R01 GM097455 and R01 CA249180 (to M.D.D.) and AstraZeneca. We thank J. Childs-Disney and R. Rahaim, Jr. for editing the manuscript. We also thank U. Bauer for supporting the collaboration and K. Jennbacken for useful input on VEGFA biology. Correspondence and request for materials should be addressed to M.D.D.
Author information
Authors and Affiliations
Contributions
M.D.D. conceived and directed the study. L.K., J.B. and M.L. conducted all physicochemical analyses and the library design for the AstraZeneca compound collection; H.S.H. conducted the screening and all in vitro and in cellulis experiments under the guidance of M.D.D.; D.A. and A.A. conducted proteomics analysis of HUVEC samples; E.L. conducted RAN translation analysis for binders to r(G4C2); K.W.W. and I.Y. conducted in silico modelling to assess the binding of the dimer to pre-miR-377; M.D.C. conducted cellular uptake analysis by LC-MS/MS; G.C. conducted all bioinformatic analyses for LOGOS analysis. All authors discussed the results and commented on the manuscript during preparation.
Corresponding author
Ethics declarations
Competing interests
M.D.D. is a founder of Expansion Therapeutics. M.L., J.B. and L.K. are employees of AstraZeneca.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–20, Discussion, Synthetic Methods and Characterization, and Supplementary Methods
Supplementary Data
Supporting Tables 1–13
Rights and permissions
About this article
Cite this article
Haniff, H.S., Knerr, L., Liu, X. et al. Design of a small molecule that stimulates vascular endothelial growth factor A enabled by screening RNA fold–small molecule interactions. Nat. Chem. 12, 952–961 (2020). https://doi.org/10.1038/s41557-020-0514-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-020-0514-4
This article is cited by
-
Anti-atherosclerotic effects of naringenin and quercetin from Folium Artemisiae argyi by attenuating Interleukin-1 beta (IL-1β)/ matrix metalloproteinase 9 (MMP9): network pharmacology-based analysis and validation
BMC Complementary Medicine and Therapies (2023)
-
Programming inactive RNA-binding small molecules into bioactive degraders
Nature (2023)