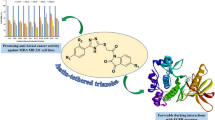
Abstract
The discovery of potent STAT3 inhibitors has gained noteworthy impetus in the last decade. In line with this trend, considering the proven biological importance of 1,2,4-triazoles, herein, we are reporting the design, synthesis, pharmacokinetic profiles, and in vitro anticancer activity of novel C3-linked 1,2,4-triazole-N-arylamide hybrids and their in silico proposed mechanism of action via inhibition of STAT3. The 1,2,4-triazole scaffold was selected as a privilege ring system that is embedded in core structures of a variety of anticancer drugs which are either in clinical use or still under clinical trials. The designed 1,2,4-triazole derivatives were synthesized by linking the triazole-thione moiety through amide hydrophilic linkers with diverse lipophilic fragments. In silico study to predict cytotoxicity of the new hybrids against different kinds of human cancer cell lines as well as the non-tumor cells was conducted. The multidrug-resistant human breast adenocarcinoma cells (MDA-MB-231) was found most susceptible to the cytotoxic effect of synthesized compounds and hence were selected to evaluate the in vitro anticancer activity. Four of the designed derivatives showed promising cytotoxicity effects against selected cancer cells, among which compound 12 showed the highest potency (IC50 = 3.61 µM), followed by 21 which displayed IC50 value of 3.93 µM. Also, compounds 14 and 23 revealed equipotent activity with the reference cytotoxic agent doxorubicin. To reinforce these observations, the obtained data of in vitro cytotoxicity have been validated in terms of ligand–protein interaction and new compounds were analyzed for ADMET properties to evaluate their potential to build up as good drug candidates. This study led us to identify two novel C3-linked 1,2,4-triazole-N-arylamide hybrids of interesting antiproliferative potentials as probable lead inhibitors of STAT3 with promising pharmacokinetic profiles.
Graphic abstract







Similar content being viewed by others
References
Jiang DM, Chan KKW, Jang RW et al (2019) Anticancer drugs approved by the Food and Drug Administration for gastrointestinal malignancies: clinical benefit and price considerations. Cancer Med 8:1584–1593. https://doi.org/10.1002/cam4.2058
Wu Q, Yang Z, Nie Y et al (2014) Multi-drug resistance in cancer chemotherapeutics: mechanisms and lab approaches. Cancer Lett 347:159–166. https://doi.org/10.1016/j.canlet.2014.03.013
Ihle JN (2001) The Stat family in cytokine signaling. Curr Opin Cell Biol 13:211–217. https://doi.org/10.1016/S0955-0674(00)00199-X
Ghosh S (2019) Cisplatin: the first metal based anticancer drug. Bioorg Chem 88:102925. https://doi.org/10.1016/j.bioorg.2019.102925
Heidelberger S, Zinzalla G, Antonow D et al (2013) Investigation of the protein alkylation sites of the STAT3:STAT3 inhibitor Stattic by mass spectrometry. Bioorg Med Chem Lett 23:4719–4722. https://doi.org/10.1016/j.bmcl.2013.05.066
Hong DS, Angelo LS, Kurzrock R (2007) Interleukin-6 and its receptor in cancer. Cancer 110:1911–1928. https://doi.org/10.1002/cncr.22999
Lee H, Deng J, Kujawski M et al (2010) STAT3-induced S1PR1 expression is crucial for persistent STAT3 activation in tumors. Nat Med 16:1421–1428. https://doi.org/10.1038/nm.2250
Sgrignani J, Garofalo M, Matkovic M et al (2018) Structural biology of STAT3 and its implications for anticancer therapies development. Int J Mol Sci 19:1591. https://doi.org/10.3390/ijms19061591
Chen H, Guan Y, Yuan G et al (2014) A perylene derivative regulates HIF-1α and Stat3 signaling pathways. Bioorg Med Chem 22:1496–1505. https://doi.org/10.1016/j.bmc.2013.10.018
Robinson AJ, Kunji ERS, Gross A (2012) Mitochondrial carrier homolog 2 (MTCH2): the recruitment and evolution of a mitochondrial carrier protein to a critical player in apoptosis. Exp Cell Res 318:1316–1323. https://doi.org/10.1016/j.yexcr.2012.01.026
Lee JH, Kim JE, Kim BG et al (2016) STAT3-induced WDR1 overexpression promotes breast cancer cell migration. Cell Signal 28:1753–1760. https://doi.org/10.1016/j.cellsig.2016.08.006
Sánchez-Ceja SG, Reyes-Maldonado E, Vázquez-Manríquez ME et al (2006) Differential expression of STAT5 and Bcl-xL, and high expression of Neu and STAT3 in non-small-cell lung carcinoma. Lung Cancer 54:163–168. https://doi.org/10.1016/j.lungcan.2006.07.012
Ke Y, Bao T, Wu X et al (2017) Scutellarin suppresses migration and invasion of human hepatocellular carcinoma by inhibiting the STAT3/Girdin/Akt activity. Biochem Biophys Res Commun 483:509–515. https://doi.org/10.1016/j.bbrc.2016.12.114
Mahanti S, Sunkara S, Bhavani R (2019) Synthesis, biological evaluation and computational studies of fused acridine containing 1,2,4-triazole derivatives as anticancer agents. Synth Commun 49:1729–1740. https://doi.org/10.1080/00397911.2019.1608450
Mioc M, Avram S, Bercean V et al (2018) Design, synthesis and biological activity evaluation of S-substituted 1H-5-Mercapto-1,2,4-triazole derivatives as antiproliferative agents in colorectal cancer. Front Chem. https://doi.org/10.3389/fchem.2018.00373
Ghanaat J, Khalilzadeh MA, Zareyee D (2020) Molecular docking studies, biological evaluation and synthesis of novel 3-mercapto-1,2,4-triazole derivatives. Mol Divers. https://doi.org/10.1007/s11030-020-10050-0
Tariq S, Kamboj P, Alam O, Amir M (2018) 1,2,4-Triazole-based benzothiazole/benzoxazole derivatives: design, synthesis, p38α MAP kinase inhibition, anti-inflammatory activity and molecular docking studies. Bioorg Chem 81:630–641. https://doi.org/10.1016/j.bioorg.2018.09.015
Bejot R, Kersemans V, Kelly C et al (2010) Pre-clinical evaluation of a 3-nitro-1,2,4-triazole analogue of [18F]FMISO as hypoxia-selective tracer for PET. Nucl Med Biol 37:565–575. https://doi.org/10.1016/j.nucmedbio.2010.03.011
Boraei ATA, Singh PK, Sechi M, Satta S (2019) Discovery of novel functionalized 1,2,4-triazoles as PARP-1 inhibitors in breast cancer: design, synthesis and antitumor activity evaluation. Eur J Med Chem 182:111621. https://doi.org/10.1016/j.ejmech.2019.111621
Kulabaş N, Tatar E, Bingöl Özakpınar Ö et al (2016) Synthesis and antiproliferative evaluation of novel 2-(4H-1,2,4-triazole-3-ylthio)acetamide derivatives as inducers of apoptosis in cancer cells. Eur J Med Chem 121:58–70. https://doi.org/10.1016/j.ejmech.2016.05.017
Ali AR, El-Bendary ER, Ghaly MA, Shehata IA (2013) Novel acetamidothiazole derivatives: synthesis and in vitro anticancer evaluation. Eur J Med Chem 69:908–919. https://doi.org/10.1016/j.ejmech.2013.08.021
Ezzat HG, Bayoumi AH, Sherbiny FF et al (2020) Design, synthesis, and molecular docking studies of new [1,2,4]triazolo[4,3-a]quinoxaline derivatives as potential A2B receptor antagonists. Mol Divers. https://doi.org/10.1007/s11030-020-10070-w
El-Morsy AM, El-Sayed MS, Abulkhair HS (2017) Synthesis, characterization and in vitro antitumor evaluation of new pyrazolo[3,4-d]pyrimidine derivatives. Open J Med Chem 07:1–17. https://doi.org/10.4236/ojmc.2017.71001
Lu X, Li X, Yang J et al (2016) Arylazolyl(azinyl)thioacetanilides. Part 20: discovery of novel purinylthioacetanilides derivatives as potent HIV-1 NNRTIs via a structure-based bioisosterism approach. Bioorg Med Chem 24:4424–4433. https://doi.org/10.1016/j.bmc.2016.07.041
El-Helby A-GA, Sakr H, Eissa IH et al (2019) Design, synthesis, molecular docking, and anticancer activity of benzoxazole derivatives as VEGFR-2 inhibitors. Arch Pharm (Weinheim). https://doi.org/10.1002/ardp.201900113
Matsuno K, Masuda Y, Uehara Y et al (2010) Identification of a new series of STAT3 inhibitors by virtual screening. ACS Med Chem Lett 1:371–375. https://doi.org/10.1021/ml1000273
Kaoud TS, Mohassab AM, Hassan HA et al (2020) NO-releasing STAT3 inhibitors suppress BRAF-mutant melanoma growth. Eur J Med Chem 186:111885. https://doi.org/10.1016/j.ejmech.2019.111885
Han J, Lim W, You D et al (2019) Chemoresistance in the human triple-negative breast cancer cell line MDA-MB-231 induced by doxorubicin gradient is associated with epigenetic alterations in histone deacetylase. J Oncol 2019:1–12. https://doi.org/10.1155/2019/1345026
Theodossiou TA, Ali M, Grigalavicius M et al (2019) Simultaneous defeat of MCF7 and MDA-MB-231 resistances by a hypericin PDT-tamoxifen hybrid therapy. NPJ Breast Cancer 5:13. https://doi.org/10.1038/s41523-019-0108-8
Ali A, Bhattacharya S (2014) DNA binders in clinical trials and chemotherapy. Bioorg Med Chem 22:4506–4521. https://doi.org/10.1016/j.bmc.2014.05.030
Abulkhair HS, Turky A, Ghiaty A et al (2020) Novel triazolophthalazine-hydrazone hybrids as potential PCAF inhibitors: design, synthesis, in vitro anticancer evaluation, apoptosis, and molecular docking studies. Bioorg Chem. https://doi.org/10.1016/j.bioorg.2020.103899
Turky A, Bayoumi AH, Ghiaty A et al (2020) Design, synthesis, and antitumor activity of novel compounds based on 1,2,4-triazolophthalazine scaffold: apoptosis-inductive and PCAF-inhibitory effects. Bioorg Chem. https://doi.org/10.1016/j.bioorg.2020.104019
Ihmaid S, Ahmed HEA, Al-Sheikh Ali A et al (2017) Rational design, synthesis, pharmacophore modeling, and docking studies for identification of novel potent DNA-PK inhibitors. Bioorg Chem 72:234–247. https://doi.org/10.1016/j.bioorg.2017.04.014
Oliveira Pedrosa M, Duarte da Cruz R, Oliveira Viana J et al (2017) hybrid compounds as direct multitarget ligands: a review. Curr Top Med Chem 17:1044–1079. https://doi.org/10.2174/1568026616666160927160620
Harrison JR, Brand S, Smith V et al (2018) A molecular hybridization approach for the design of potent, highly selective, and brain-penetrant N-myristoyltransferase inhibitors. J Med Chem 61:8374–8389. https://doi.org/10.1021/acs.jmedchem.8b00884
Bayoumi A, Ghiaty A, El-Morsy A et al (2012) Synthesis and evaluation of some new 1,2,4-triazolo(4,3-a)quinoxalin-4-5H-one derivatives as AMPA receptor antagonists. Bull Fac Pharmacy, Cairo Univ 50:141–146. https://doi.org/10.1016/j.bfopcu.2012.05.002
Pagadala R, Meshram JS, Chopde HN et al (2012) Synthesis and antimicrobial evaluation of new monocyclic β-lactams. J Heterocycl Chem 49:1151–1155. https://doi.org/10.1002/jhet.973
Abul-Khair H, Elmeligie S, Bayoumi A et al (2013) Synthesis and evaluation of some new (1,2,4) Triazolo(4,3-a)Quinoxalin-4(5H)-one derivatives as AMPA receptor antagonists. J Heterocycl Chem 50:1202–1208. https://doi.org/10.1002/jhet.714
El-Helby AA, Ayyad RRA, Zayed MF et al (2019) Design, synthesis, in silico ADMET profile and GABA—a docking of novel phthalazines as potent anticonvulsants. Arch Pharm (Weinheim) 352:1800387. https://doi.org/10.1002/ardp.201800387
Eid I, Elsebaei MM, Mohammad H et al (2017) Arylthiazole antibiotics targeting intracellular methicillin-resistant Staphylococcus aureus (MRSA) that interfere with bacterial cell wall synthesis. Eur J Med Chem 139:665–673. https://doi.org/10.1016/j.ejmech.2017.08.039
Hannoun MH, Hagras M, Kotb A et al (2020) Synthesis and antibacterial evaluation of a novel library of 2-(thiazol-5-yl)-1,3,4-oxadiazole derivatives against methicillin-resistant Staphylococcus aureus (MRSA). Bioorg Chem. https://doi.org/10.1016/j.bioorg.2019.103364
Omar AM, Ihmaid S, Habib E-SE et al (2020) The rational design, synthesis, and antimicrobial investigation of 2-Amino-4-Methylthiazole analogues inhibitors of GlcN-6-P synthase. Bioorg Chem. https://doi.org/10.1016/j.bioorg.2020.103781
Lagunin AA, Dubovskaja VI, Rudik AV et al (2018) CLC-Pred: a freely available web-service for in silico prediction of human cell line cytotoxicity for drug-like compounds. PLoS ONE 13:e0191838. https://doi.org/10.1371/journal.pone.0191838
Morgan DML (ed) (1998) Tetrazolium (MTT) Assay for cellular viability and activity. In: Polyamine protocols. Methods in molecular biology™, vol 79. Humana Press. https://doi.org/10.1385/0-89603-448-8:179
Bai L, Zhou H, Xu R et al (2019) A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell 36:498–511.e17. https://doi.org/10.1016/j.ccell.2019.10.002
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23:3–25. https://doi.org/10.1016/S0169-409X(96)00423-1
DE Pires V, Blundell TL, Ascher DB (2015) pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 58:4066–4072. https://doi.org/10.1021/acs.jmedchem.5b00104
Beig A, Agbaria R, Dahan A (2013) Oral delivery of lipophilic drugs: the tradeoff between solubility increase and permeability decrease when using cyclodextrin-based formulations. PLoS ONE 8:e68237. https://doi.org/10.1371/journal.pone.0068237
Murty MSR, Ram KR, Venkateswara Rao R et al (2012) Synthesis of new S-alkylated-3-mercapto-1,2,4-triazole derivatives bearing cyclic amine moiety as potent anticancer agents. Lett Drug Des Discov 9:276–281. https://doi.org/10.2174/157018012799129882
Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Turky, A., Bayoumi, A.H., Sherbiny, F.F. et al. Unravelling the anticancer potency of 1,2,4-triazole-N-arylamide hybrids through inhibition of STAT3: synthesis and in silico mechanistic studies. Mol Divers 25, 403–420 (2021). https://doi.org/10.1007/s11030-020-10131-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-020-10131-0