Abstract
Purpose
Health-care practitioners’ (HCPs) preferences for returning secondary findings (SFs) will influence guideline compliance, shared decision-making, and patient health outcomes. This study aimed to estimate HCPs’ preferences and willingness to support the return (WTSR) of SFs in Canada.
Methods
A discrete choice experiment estimated HCPs’ preferences for the following attributes: disease risk, clinical utility, health consequences, prior experience, and patient preference. We analyzed responses with an error component mixed logit model and predicted WTSR using scenario analyses.
Results
Two hundred fifty participants of 583 completed the questionnaire (completion rate: 42.9%). WTSR was significantly influenced by patient preference and SF outcome characteristics. HCPs’ WTSR was 78% (95% confidence interval: 74–81%) when returning SFs with available medical treatment, high penetrance, severe health consequences, and patient’s preference for return. Genetics professionals had a higher WTSR than HCPs of other types when returning SFs with clinical utility and patient preference to know. HCPs >55 years of age were more likely to return SFs compared with younger HCPs.
Conclusion
This study identified factors that influence WTSR of SFs and indicates that HCPs make tradeoffs between patient preference and other outcome characteristics. The results can inform clinical scenarios and models aiming to understand shared decision-making, patient and family opportunity to benefit, and cost-effectiveness.
Similar content being viewed by others
INTRODUCTION
Next-generation sequencing (NGS) clinical tests provide information about disease risk, prognosis, or treatment response.1,2 In some instances, NGS may identify variants that are unrelated to the primary intention for testing, which are called secondary findings (SFs).3,4,5 The return of SFs from genome-scale sequencing utilizing NGS is debated.6,7,8,9 On one hand, some pathogenic variants are not medically actionable, and the disclosure of these variants may cause patient anxiety and psychological harm,10 or induce information overload.11,12 On the other hand, nondisclosure of SFs may miss the chance of informing patients of life-threatening risks; hence patients may miss interventional recommendations and life-planning options to mitigate the risks.13,14,15
Clinical guidelines on genome-scale sequencing suggest that physicians should return SFs with clinical utility (e.g., medical actionability) and high penetrance.4,16 The American College of Medical Genetics and Genomics (ACMG) has published a list of gene–disease pairs (currently consisting of 59 genes) that should be returned to patients on a consent basis.5 The list is expected to evolve as the evidence base on penetrance and medical actionability increases. In contrast, guidelines from the Canadian College of Medical Geneticists (CCMG) take a cautious approach and recommend avoiding the clinical return of SFs, although patients can request SF detection.3 The disparity in clinical guidelines reflects a lack of evidence on the clinical, cost, and psychosocial consequences of returning SFs, and insufficient incorporation of patient preference and physicians' views in guidelines.17
Previous studies have provided evidence of patients’ preferences for the return of SFs, indicating most individuals valued the receipt of SFs with health benefits depending on the penetrance, medical actionability, and the cost of obtaining the findings.18,19 These studies are important to support informed decision-making,18,20 but there is limited quantitative evidence surrounding health-care practitioners’ (HCPs) preferences for the return of SFs,21 particularly in terms of willingness to support the disclosure of genomic results. This evidence is important because HCPs may act as gatekeepers when returning information on SFs. Their willingness to disclose results will have implications for shared decision-making, patient and family opportunity to benefit, clinical effectiveness, and cost-effectiveness.22 For example, in situations where SFs with medical actionability and high penetrance are detected, HCPs’ willingness to disclose SFs will influence the number of individuals and families that benefit from SF information. The more individuals that benefit from SF information with effective clinical management, the more likely that returning SFs improves population health and is cost-effective. Current studies have qualitatively identified several important factors when HCPs consider the return of SFs,21,23,24 including patient consent, clinical utility, analytical validity, test accuracy, age of onset, disease severity, and chance of developing diseases. A published discrete choice experiment (DCE) examined Australian genetics professionals’ preferences toward the return of SFs, using the attributes of chance of developing a disorder, age of onset, disease severity, and availability of prevention and treatment.25 However, the published DCE did not include patient preference as an attribute, and did not examine the predicted willingness of HCPs to support the return of SFs (WTSR). This omission is significant because the revised ACMG policy recommendations have stated that patients should be allowed to opt out of receiving SFs. The influence of patient preference on HCPs’ WTSR of SFs has not been examined, however. The objective of this study is to estimate HCPs’ preferences for returning SFs, and to estimate HCPs’ WTSR of SFs in the context of different policy scenarios.
MATERIALS AND METHODS
Ethics statement
The University of British Columbia Behavioral Research Ethics Board (UBC BREB) at BC Cancer approved the study (H15-02492). Informed consent was obtained from subjects prior to survey initiation. Data collection and analysis was performed in accordance with UBC BREB regulations.
Discrete choice experiment
We conducted a survey among HCPs, in which a DCE elicited HCPs’ preferences and WTSR of SFs. The DCE method is an established stated preference approach to simulate the effect that the factors of a good or service have on individual choice.26 The DCE method generates choice scenarios through tasks that require respondents to select one alternative among several, designed to mimic real-world decision-making. In stating a preference, the respondent is assumed to choose the alternative that yields the highest benefit.27
A DCE starts by first identifying the characteristics (called attributes) of a good or service. Attributes are defined across a range of levels that describe the good or service. Experimental design techniques are applied to construct a series of choice tasks, each composed of hypothetical scenarios defined by combinations of attribute levels.26 When a respondent selects between scenarios, they are making tradeoffs. Analysis of the responses provides parameter estimates of preference-based value from which tradeoffs between attributes can be calculated.27 The parameter estimates can further be used to estimate HCPs’ WTSR of SFs.
Questionnaire development
The attributes were initially identified by a scoping review of the literature.18,24,25,28 The results informed the creation of the attributes and levels of a pilot DCE. We recruited a convenience sample of HCPs with experience returning genomic results (N = 9)25 to collect their opinions on the pilot DCE. The sample provided written feedback and we revised the attributes and associated levels accordingly.
In total, we included five attributes: risk for developing a health condition at some point in the future, clinical utility (e.g., medical actionability) of the finding, health consequences of the condition, prior experience in managing the health condition, and patient’s preference for the return of information (Table 1). For future applicability, we took the recommendations by ACMG guidelines and incorporated the possibility of pharmacogenomic variants indicated by SFs in the clinical utility attribute,2,5 by specifying one level as “a list of medications that are highly likely to be more effective or cause side effects is available.”
In each choice task, respondents were required to choose the profile (i.e., a set of SFs awaiting disclosure decision) in which they were most comfortable returning SFs (Fig. 1). Choice tasks included two hypothetical profiles, each of which was composed by levels from the five attributes. A “Neither” option was included into each choice task to account for the possibility that HCPs did not support the return of SFs.16 The DCE experimental design creating the choice tasks was generated using a D-optimal approach.29 This experimental design included 48 choice tasks, which were divided into 3 blocks such that each participant faced 16 choices. Included in the DCE survey were five demographic questions comprising age, gender, professional filed, practicing years, location, and frequency of ordering genomic tests or interpreting results (i.e., experiential characteristics). The questionnaires were available in English or French.
Population and sample
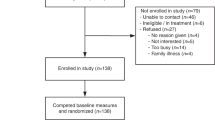
The study population was Canadian HCPs. We recruited participants through the assistance of CCMG, the Canadian Association of Genetic Counselors (CAGC), and the Royal College of Physicians and Surgeons of Canada. We sent invitation emails to interested members of these professional organizations and reminded them later by two follow-up emails at two-week intervals to facilitate participation. Inclusion criteria were HCPs eligible to order genomic tests or interpretation of results. Once HCPs consented to participate, each participant was referred to a password-protected website where our study questionnaire was included.
Statistical analysis
Data were analyzed in STATA 14.2 (StataCorp LP) using an error components mixed logit model.30 The categorical attributes were effects-coded. Reference levels were specified for each attribute. The parameters of attribute levels were assumed to follow the normal distribution. Estimations of parameters are relative to the reference level within each attribute. The mean of a parameter represents the average preference value that respondents associate with the attribute level, and the standard deviation (SD) characterizes the heterogeneity of the preference value among respondents. By applying the mean and SD of a preference value associated with an attribute level, we calculated the probability that the value was less than 0, which indicated the percentage of HCPs who would have a negative preference value for returning the type of SFs as defined by the attribute level.
All potential interactions between the characteristics of respondents and attribute levels were examined. The interaction terms were selected using a backward selection method, based on the contribution of each term to model fit. The log-likelihood ratio test was employed to compare the model specifications and a reduced model with one interaction term removed. If the removed term influenced the model fit significantly, the term was retained. We further conduct split sample analyses of genetic counselors and medical geneticists (see Supplementary Materials and Methods).
We used the nonreturn of SFs as the base policy scenario, which is also the prevailing policy in Canada and in other jurisdictions.3,31,32 Given the ACMG recommended the disclosure of SFs associated with high penetrance, severe health consequences, medical treatment, and patient’s preference to receive actionable SFs,4,5 we constructed several policy scenarios following these recommendations as contrast to the nonreturn scenario to estimate HCPs’ WTSR. The WTSR indicated the percentage of HCPs predicted to support the new policy scenario compared with nonreturn.26 We used the model parameters to estimate the WTSR values, following the closed-form formula for choice probability calculation.30 The 95% confidence intervals (CIs) for WTSR were generated using delta method.18
RESULTS
Respondent characteristics
In total, 583 HCPs responded to the email invitation by indicating acceptance or refusal, of whom 250 completed the questionnaire (completion rate 42.9%). Table 2 presents an overview of participants’ characteristics. The majority of respondents (51.2%) were genetics professionals, among whom genetic counselors (38.4%) were more prevalent than medical geneticists (12.8%). The respondent cohort had a mixed level of familiarity with ordering genomic tests or interpreting results, with a majority (52.8%) indicating that they are “always” or “often” involved in such activities. Participants were predominantly based in Ontario (58%) with representation across the regions in Canada.
Factors influencing the return of SFs
The estimated regression parameters and the relative importance of the attributes are in Fig. 2 and reported in the Supplementary Material and Methods Table S1. Respondents were more willing to return SFs with moderate or high penetrance (lifetime disease risk ≥40% or ≥80%), with severe or very severe health consequences, and when patients preferred to know the results. The availability of recommended medical treatment, lifestyle intervention, and pharmacogenomic information also positively influenced the WTSR of SFs, as did prior experience in disease management. Respondents were less likely to return SFs associated with low penetrance disorders (lifetime disease risk ≥5% or ≥25%), with mild or moderate health consequences, and when the patient preferred not to know about SFs. The lack of effective intervention also made HCPs less likely to return SFs.
The mean relative importance score was calculated by computing the difference between the highest and lowest rescaled coefficients for the levels of that attribute. Mean relative importance represents change in willingness to support the return of results from the lowest attribute level to the highest, expressed on a scale from 0 to 100. Med refers to the clinical utility level of “recommended effective medical treatment is available.” PGx treatments based on pharmacogenomic variants indicated by secondary findings (SFs), referring to the level of “list of medications that are highly likely to be more effective or cause side effects is available.” Lifestyle refers to the level of “recommended effective lifestyle or behavior modification is available.” None refers to the level of “no effective medical treatment or lifestyle changes recommended.” Accept all SFs refers to the patient preference level of “patient wants to know all findings regardless of clinical utility.” Accept SFs with CU refers to the level of “patient only wants to know findings with clinical utility”.
Results indicated that, among all attribute levels and all else equal, “patient wants to know SFs regardless of clinical utility” and “patient does not want to know SFs” generated the largest (mean = 1.36) and smallest (mean = -2.28) relative preference values for HCPs, implying that they were factors that made HCPs most and least willing to return SFs, respectively. Further, among available interventional options, treatments based on pharmacogenomic information (mean = 0.56) generated higher preference value (p < 0.05) than lifestyle modification (mean = 0.29).
As illustrated in Fig. 2, the preference values within three attributes (i.e., penetrance, clinical utility, and patient preference) had a categorically ordered increase or decrease across levels. For example, the preference value generated by low penetrance (mean = -0.10) was smaller than that generated by moderate penetrance (mean = 0.20); moderate penetrance generated a smaller preference value than high penetrance did (mean = 1.00). HCPs had a slightly higher preference value (p < 0.05) for returning SFs associated with “severe health consequences” (mean = 0.66) than that with “very severe health consequences” (mean = 0.64), a result observed by Regier et al.18 in context to public preferences for the return of SFs. HCPs with “some experience” (mean = 0.39) or “moderate experience” (mean = 0.21) treating diseases indicated by the SFs were more willing (p < 0.05) to return findings than HCPs with “substantial experience” (mean = 0.15).
Interaction with demographic characteristics
The interaction model contained seven statistically significant interaction terms (Table S2 in Supplementary Materials and Methods). The model results indicated that genetics professionals (i.e., medical geneticists and genetic counselors) were more willing to return SFs than other HCPs (e.g., cardiologist, oncologist), when patients preferred to learn SFs with clinical utility. Genetics professionals were also more willing to return SFs than HCPs of other types when recommended medical treatments were available for health conditions indicated by the SFs. In addition, older HCPs (≥55 years) were predicted to be more willing to return SFs compared with younger ones. The interactions between the frequency of ordering or interpreting genomic tests and attribute levels were insignificant, suggesting the frequency had no significant impact on WTSR.
Scenario analysis of willingness to return secondary findings
Scenario 1 (Table 3) aligns with the ACMG working group recommendations (disorders with recommended medical treatment, high penetrance, severe health consequences, and patient’s preference to receive clinically actionable SFs),4,5 where the analysis predicted that 78% (95% CI: 74–81%) of HCPs would be willing to return SFs. Scenario 2 aligns with the first scenario in SF outcome characteristics but without information on patient preference (i.e., recommended medical treatment available, high penetrance, and severe health consequences), where we predicted 74% (95% CI: 70–78%) of HCPs would be willing to return the SFs. Scenario 3 examined the WTSR for SFs with moderate penetrance (≥40%), severe health consequences, and available medical treatment. It is predicted that under this scenario, 72% (95% CI: 68–76%) of HCPs would return the SFs. Scenarios 4 and 5 describe disorders with severe health consequences where pharmacogenomic information for clinical interventions are available. Among high penetrance conditions (scenario 4), the model predicted that 69% (95% CI: 65–73%) of HCPs would prefer to return the SFs. The predicted WTSR was 67% (95% CI: 62–71%) among conditions with a moderate penetrance (scenario 5).
Returning SFs despite patient preference not to know
We found that patient preference was not an absolute factor in determining the WTSR of HCPs. Instead, HCPs made tradeoffs between patient preference and the characteristics of SFs (i.e., penetrance, clinical utility, and health consequences) when asked about their preferences to return SFs. We identified three scenarios in which a majority of HCPs were willing to disclose SFs when the patient preferred not to know (scenario analysis results are in Table S3). In the first scenario (scenario S3.1 in Table S3), we predicted that 55% (95% CI: 49–60%) of HCPs would be willing to return SFs with high penetrance (≥80%), very severe health consequences, and effective medical treatment available when the patient did not want to know. If health consequences were severe while the other two characteristics remained as per the first scenario (i.e., high penetrance and effective medical treatment available), we predicted 54% (95% CI: 48–59%) of HCPs would be willing to return SFs (scenario S3.2). If the penetrance decreased to moderate level (≥40%) and other characteristics remained as per the first scenario (i.e., very severe health consequences and effective medical treatment available), we predicted 52% (95% CI: 46–58%) of HCPs would be willing to return SFs (scenario S3.3).
No return of SFs despite patient preference to know
The scenario analysis identified six scenarios in which a majority of HCPs were not willing to return SFs when patients said they preferred to know about SFs (Table S4), again confirming that HCPs made tradeoffs between patient preference and the characteristics of SFs. Scenario S4.1 predicted that 70% (95% CI: 65–75%) of HCPs would not return SFs with very low (≥5%) penetrance, no treatment available, and mild health consequences. Even when the health consequences increased to moderate, severe and very severe levels (scenarios S4.2, S4.3, and S4.4, respectively), a majority of HCPs (69%, 56%, and 55%, respectively) favored nonreturn despite patient preference. The last two scenarios predicted that around 60% of HCPs would not return SFs with low penetrance (≥25%), no treatment available, and mild or moderate health consequences (scenarios S4.5 and S4.6, respectively), despite patient’s preference to know.
DISCUSSION
This study quantified the preferences of HCPs on the return of SFs derived from clinical genome-scale sequencing and predicted their willingness to return SFs in policy-relevant scenarios. HCPs’ willingness to return was highly influenced by the patient preference to receive SFs and by the characteristics of SFs (i.e., penetrance, clinical utility, and health consequences), without much consideration of their own prior experience in managing the disorder indicated by SFs. Among all the attribute levels, “patient wants to know SFs regardless of clinical utility” and “patient does not want to know SFs” emerged as significant factors that made HCPs most and least willing to return SFs, all else equal. HCPs made tradeoffs between patient preference and the characteristics of SFs when asked about their preferences in returning SFs. Additionally, we found that genetics professionals and older HCPs were more willing to return SFs than other HCPs were, indicating that practicing fields and age also affected HCPs’ WTSR.
We found that patient preference was not absolute in determining HCPs’ WTSR. We identified policy scenarios where HCPs would be willing (or unwilling) to return SFs, irrespective of patient preference. Our scenario analysis suggests that HCPs would return SFs in three scenarios associated with moderate or high penetrance, medical treatment available, and severe or very severe health consequences, despite that the patient preferred not to know SFs. Conversely, we identified six scenarios associated with low penetrance, no treatment available, and different levels of health consequences, where HCPs would not return SFs despite patient preference to know. These scenarios can be explicitly considered by all relevant stakeholders in debate that may inform future amendments to the guidelines.
Our analysis found that HCPs’ WTSR differs between professional types. Genetics professionals were more comfortable to disclose SFs compared with HCPs of other types. Greater familiarity and expertise with genetic technologies may be a factor in overall WTSR of genomic information to patients. We encourage future analyses to examine HCPs’ preferences considering various familiarity level and expertise with genetic technologies. Further, this result may suggest that decision tools on ordering genomic tests and returning findings would be helpful for HCPs who are less familiar with these technologies, as these tools have been shown to be effective in improving patient knowledge while reducing time spent with HCPs thus streamlining educational efforts and counseling expertise.33,34
This article also presents evidence that HCPs make tradeoffs between their clinical experience and the characteristics of SFs. HCPs are willing to return SFs in some policy scenarios even when they have “no experience” in managing the disorder indicated by the SFs. It is worth noting that “some experience” generated higher preference value than “moderate experience” and “substantial experience” did. The result was unexpected. It is possible that respondents did not consistently distinguish between “some,” “moderate,” and “substantial” experience when completing choice tasks. It may be the case that those with substantial experience with managing a disease feel that returning SFs can alter health outcomes. Finally, the choice task presented was complex. Task complexity has been shown to negatively influence cognitive processing in DCEs.35
Cost-effectiveness analyses (CEA) are recommended for informing health-care policy in different countries (United States, Canada, UK, etc.).36,37,38 CEAs that examine the value of genome-scale sequencing face methodological challenges that remain to be addressed, including the incorporation of returning SFs derived from genome-scale sequencing and associated impacts.39 One existing study in the literature provided an initial approach to evaluate the economic outcomes of returning SFs.22 Future CEAs measuring the value of genome-scale sequencing could consider incorporating the return of SFs and develop approaches to consider the downstream impacts of returning SFs. For example, if a physician wishes to return SFs and the patient wishes to receive them, the disclosure of results may lead to a change in clinical management (e.g., intensive surveillance or prophylactic surgery) and an increase in management costs.40 To better capture the impacts of returning SFs on the costs, it is important to identify policy scenarios in which SFs are returned with the consideration of both patient and practitioner preferences. A shared decision-making approach requires that both perspectives be accommodated.18,20 Our analysis provides estimates of how HCPs’ preferences may influence the return of genomic findings through the establishment of specific policy scenarios, and invites researchers and decision-makers to consider the specific scenarios where clinical management changes may occur.
Limitations
Our study has several limitations. First, our sample was limited to a total of 250 HCPs within Canada with a 42.9% response rate. While the response rate is similar to other studies,18 it will likely not be broadly representative, especially for the nonresponders. We were unable to assess if responders were significantly different from nonresponders. Second, the DCE required the HCPs to select choices among hypothetical scenarios. The estimates reflected their stated preference, which might be different from how they would act in the real world. The hypothetical nature of DCEs may impede the HCPs’ consideration of all realistic constraints (i.e., “hypothetical bias”). Third, although the attributes were determined deliberately through literature review and discussion with experts, there may be some factors not included in the attributes. In this case, we cannot measure the tradeoffs associated with those unidentified factors. Fourth, the interpretation of the attribute levels may differ between respondents. For example, the attribute level of “medications highly likely to be more effective or cause side effects” may not be interpreted as information related to pharmacogenomics by all respondents. Finally, there might be sampling bias in the DCE because the respondents were invited to complete the survey and not randomly selected. We cannot assess whether our sample is representative of the population of eligible HCPs in Canada. These limitations challenge the generalizability of our results.
Conclusion
Our study predicting the willingness of HCPs to return SFs has important policy implications. First, our study suggests the disclosure of SFs by HCPs is significantly influenced by patient preference. We present evidence on the types of scenarios in which HCPs are inclined to return SFs (e.g., SFs associated with high penetrance, severe health consequences, and recommended medical treatment). We also describe scenarios where current ACMG guidelines may or may not be adhered to and when patient preference for return of results are respected. For example, HCPs are reluctant to return SFs associated with low penetrance, no treatment available, and mild health consequences, despite patient preference to know. These identified scenarios may warrant further stakeholder debate around SFs and suggest the value of developing clinical decision tools for HCPs when ordering genomic tests and returning results.
Second, the CCMG guidelines cite a lack of health economic evidence as an obstacle to the creation of Canadian guidelines for the return of SFs.3 This work provides important context to the discussion on the costs and effectiveness of SF disclosure. If SFs are returned, this will influence both downstream costs and effectiveness predicted by the economic models. Future CEAs of genome-scale sequencing can utilize the estimates of this study to refine the economic models and more accurately capture the downstream impacts of returning SFs, generating evidences for future guidelines.
References
Biesecker LG. Opportunities and challenges for the integration of massively parallel genomic sequencing into clinical practice: lessons from the ClinSeq project. Genet Med. 2012;14:393–398.
Green RC, Rehm HL, Kohane IS. Clinical genome sequencing. Genom Pers Med. 2013;2:102–122.
Boycott K, Hartley T, Adam S, et al. The clinical application of genome-wide sequencing for monogenic diseases in Canada: position statement of the Canadian College of Medical Geneticists. J Med Genet. 2015;52:431–437.
Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565.
Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2. 0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249.
Grove ME, Wolpert MN, Cho MK, Lee SS-J, Ormond KE. Views of genetics health professionals on the return of genomic results. J Genet Couns. 2014;23:531–538.
Strong K, Zusevics K, Bick D, Veith R. Views of primary care providers regarding the return of genome sequencing incidental findings. Clin Genet. 2014;86:461–468.
Yu J-H, Harrell TM, Jamal SM, Tabor HK, Bamshad MJ. Attitudes of genetics professionals toward the return of incidental results from exome and whole-genome sequencing. Am J Hum Genet. 2014;95:77–84.
Scheuner MT, Peredo J, Benkendorf J, et al. Reporting genomic secondary findings: ACMG members weigh. Genet Med. 2015;17:27.
Wolf SM, Lawrenz FP, Nelson CA, et al. Managing incidental findings in human subjects research: analysis and recommendations. J Law Med Ethics. 2008;36:219–248.
Cho MK. Understanding incidental findings in the context of genetics and genomics. J Law Med Ethics. 2008;36:280–285.
Hallowell N, Hall A, Alberg C, Zimmern R. Revealing the results of whole-genome sequencing and whole-exome sequencing in research and clinical investigations: some ethical issues. J Med Ethics. 2015;41:317–321.
Caulfield T, McGuire AL, Cho M, et al. Research ethics recommendations for whole-genome research: consensus statement. PLoS Biol. 2008;6:e73.
McGuire AL, Caulfield T, Cho MK. Research ethics and the challenge of whole-genome sequencing. Nat Rev Genet. 2008;9:152.
Hart MR, Biesecker BB, Blout CL, et al. Secondary findings from clinical genomic sequencing: prevalence, patient perspectives, family history assessment, and health-care costs from a multisite study. Genet Med. 2019;21:1100.
ACMG Board of Directors. ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med. 2015;17:68–69.
Shickh S, Clausen M, Mighton C, et al. Health outcomes, utility and costs of returning incidental results from genomic sequencing in a Canadian cancer population: protocol for a mixed-methods randomised controlled trial. BMJ Open. 2019;9:e031092–e031092.
Regier DA, Peacock SJ, Pataky R, et al. Societal preferences for the return of incidental findings from clinical genomic sequencing: a discrete-choice experiment. CMAJ. 2015;187:E190–E197.
Ploug T, Holm S. Clinical genome sequencing and population preferences for information about ‘incidental’findings'—from medically actionable genes (MAGs) to patient actionable genes (PAGs). PLoS One. 2017;12:e0179935.
Barry MJ, Edgman-Levitan S. Shared decision making—the pinnacle of patient-centered care. N Engl J Med. 2012;366:780–781.
Lohn Z, Adam S, Birch P, Townsend A, Friedman J. Genetics professionals’ perspectives on reporting incidental findings from clinical genome‐wide sequencing. Am J Med Genet A. 2013;161:542–549.
Bennette CS, Gallego CJ, Burke W, Jarvik GP, Veenstra DL. The cost-effectiveness of returning incidental findings from next-generation genomic sequencing. Genet Med. 2014;17:587–595.
Wolf SM, Annas GJ, Elias S. Respecting patient autonomy in clinical genomics: new recommendations on incidental findings go astray. Science. 2013;340:1049.
Turbitt E, Halliday JL, Metcalfe SA. Key informants’ perspectives of implementing chromosomal microarrays into clinical practice in Australia. Twin Res Hum Genet. 2013;16:833–839.
Turbitt E, Wiest MM, Halliday JL, Amor DJ, Metcalfe SA. Availability of treatment drives decisions of genetic health professionals about disclosure of incidental findings. Eur J Hum Genet. 2014;22:1225–1228.
Ryan M Gerard K Amaya-Amaya MUsing discrete choice experiments to value health and health care. Dordrecht, Netherlands: Springer; 2007.
McFadden D Conditional logit analysis of qualitative choice behavior. In: Frontiers in Econometrics. Zarembka P, editor. Wiley, New York: 1973.
Bennette CS, Trinidad SB, Fullerton SM, et al. Return of incidental findings in genomic medicine: measuring what patients value - development of an instrument to measure preferences for information from next-generation testing (IMPRINT). Genet Med. 2013;15:873–881.
Johnson FR, Lancsar E, Marshall D, et al. Constructing experimental designs for discrete-choice experiments: report of the ISPOR conjoint analysis experimental design good research practices task force. Value Health. 2013;16:3–13.
Train KE. Discrete choice methods with simulation. Cambridge: Cambridge University Press; 2009.
Van ElCG, Cornel MC, Borry P, et al. Whole-genome sequencing in health care: recommendations of the European Society of Human Genetics. Eur J Hum Genet. 2013;21:580.
Hall A, Finnegan T, Alberg C. Realising genomics in clinical practice. Cambridge: PHG Foundation; 2014.
Bombard Y, Clausen M, Shickh S, et al. Effectiveness of the Genomics ADvISER decision aid for the selection of secondary findings from genomic sequencing: a randomized clinical trial. Genet Med. 2019;22:727–735.
Mighton C, Carlsson L, Clausen M, et al. Development of patient “profiles” to tailor counseling for incidental genomic sequencing results. Eur J Hum Genet. 2019;27:1008–1017.
Regier DA, Watson V, Burnett H, Ungar WJ. Task complexity and response certainty in discrete choice experiments: an application to drug treatments for juvenile idiopathic arthritis. J Behav Exp Econ. 2014;50:40–49.
CADTH. Guidelines for the economic evaluation of health technologies: Canada. In. 4th ed. Ottawa, Canada: CADTH; 2017.
Sanders GD, Neumann PJ, Basu A, et al. Recommendations for conduct, methodological practices, and reporting of cost-effectiveness analyses: second panel on cost-effectiveness in health and medicine. JAMA. 2016;316:1093–1103.
National Institute for Health and Care Excellence. Assessing cost effectiveness. London, United Kingdom: National Institute for Health and Care Excellence; 2012.
Phillips KA, Deverka PA, Marshall DA, et al. Methodological issues in assessing the economic value of next-generation sequencing tests: many challenges and not enough solutions. Value Health. 2018;21:1033–1042.
Linley WG, Hughes DA. Decision-makers’ preferences for approving new medicines in Wales: a discrete-choice experiment with assessment of external validity. Pharmacoeconomics. 2013;31:345–355.
Acknowledgements
This research was funded by the Canadian Centre for Applied Research in Cancer Control (ARCC). ARCC is funded by the Canadian Cancer Society (2015-703549). Y.B. was supported by a Canadian Institute of Health Research New Investigator Award during this study. We thank all participants who took part in the survey.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
D.A.R. reports conference travel funding from Illumina. The other authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Jiang, S., Anis, A.H., Cromwell, I. et al. Health-care practitioners’ preferences for the return of secondary findings from next-generation sequencing: a discrete choice experiment. Genet Med 22, 2011–2019 (2020). https://doi.org/10.1038/s41436-020-0927-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-0927-x
Keywords
This article is cited by
-
Eliciting parental preferences and values for the return of additional findings from genomic sequencing
npj Genomic Medicine (2024)
-
Gauging Incentive Values and Expectations (G.I.V.E.) among Blood Donors for Nonmonetary Incentives: Developing a Preference Elicitation Instrument through Qualitative Approaches in Shandong, China
The Patient - Patient-Centered Outcomes Research (2023)
-
Patient Preferences in Targeted Pharmacotherapy for Cancers: A Systematic Review of Discrete Choice Experiments
PharmacoEconomics (2023)
-
Breast Cancer Screening Should Embrace Precision Medicine: Evidence by Reviewing Economic Evaluations in China
Advances in Therapy (2023)