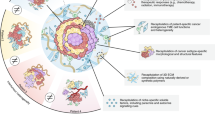
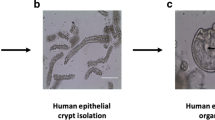
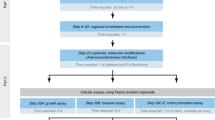
Abstract
Organoid technologies enable the creation of in vitro physiologic systems that model tissues of origin more accurately than classical culture approaches. Seminal characteristics of these systems, including three-dimensional structure and recapitulation of self-renewal, differentiation and disease pathology, render organoids eminently suited as hybrids that combine the experimental tractability of traditional two-dimensional cell lines with cellular attributes of in vivo model systems. Here we describe recent advances in this rapidly evolving field and their application to cancer biology, clinical translation and precision medicine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Begley, C. G. & Ellis, L. M. Drug development: raise standards for preclinical cancer research. Nature 483, 531–533 (2012).
Lancaster, M. A. & Knoblich, J. A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125 (2014).
Lancaster, M. A. & Huch, M. Disease modelling in human organoids. Dis. Model. Mech. 12, dmm039347 (2019).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Ootani, A. et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 15, 701–706 (2009).
Spence, J. R. et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109 (2011).
Fujii, M. et al. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell 23, 787–793.e6 (2018).
Jung, P. et al. Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 17, 1225–1227 (2011).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011).
Barker, N. et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 (2010).
Bartfeld, S. et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148, 126–136.e6 (2015).
DeWard, A. D., Cramer, J. & Lagasse, E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Reports 9, 701–711 (2014).
Maimets, M. et al. Long-term in vitro expansion of salivary gland stem cells driven by Wnt signals. Stem Cell Reports 6, 150–162 (2016).
Nanduri, L. S. Y. et al. Purification and ex vivo expansion of fully functional salivary gland stem cells. Stem Cell Reports 3, 957–964 (2014).
Ren, W. et al. Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc. Natl. Acad. Sci. USA 111, 16401–16406 (2014).
Boj, S. F. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015).
Georgakopoulos, N. et al. Long-term expansion, genomic stability and in vivo safety of adult human pancreas organoids. BMC Dev. Biol. 20, 4 (2020).
Wang, D. et al. Long-term expansion of pancreatic islet organoids from resident Procr+ progenitors. Cell 180, 1198–1211.e19 (2020).
Loomans, C. J. M. et al. Expansion of adult human pancreatic tissue yields organoids harboring progenitor cells with endocrine differentiation potential. Stem Cell Reports 10, 712–724 (2018).
Huch, M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013).
Hu, H. et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 175, 1591–1606.e19 (2018).
Huch, M. et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312 (2015).
Peng, W. C. et al. Inflammatory cytokine TNFα promotes the long-term expansion of primary hepatocytes in 3D culture. Cell 175, 1607–1619 (2018).
Sachs, N. et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 38, e100300 (2019).
Danahay, H. et al. Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Reports 10, 239–252 (2015).
Tata, P. R. et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 503, 218–223 (2013).
Rock, J. R. et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 106, 12771–12775 (2009).
Sachs, N. et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–386.e10 (2018).
Chua, C. W. et al. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat. Cell Biol. 16, 951–961 (2014).
Karthaus, W. R. et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159, 163–175 (2014).
Schutgens, F. et al. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat. Biotechnol. 37, 303–313 (2019).
Kessler, M. et al. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat. Commun. 6, 8989 (2015).
Kopper, O. et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 25, 838–849 (2019).
Hill, S. J. et al. Prediction of DNA repair inhibitor response in short-term patient-derived ovarian cancer organoids. Cancer Discov. 8, 1404–1421 (2018).
Turco, M. Y. et al. Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat. Cell Biol. 19, 568–577 (2017).
Boonekamp, K. E. et al. Long-term expansion and differentiation of adult murine epidermal stem cells in 3D organoid cultures. Proc. Natl. Acad. Sci. USA 116, 14630–14638 (2019).
Bhaduri, A. et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature 578, 142–148 (2020).
Ganesh, K. et al. L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer. Nat. Cancer 1, 28–45 (2020).
Co, J. Y. et al. Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Reports 26, 2509–2520 (2019).
Neal, J. T. et al. Organoid modeling of the tumor immune microenvironment. Cell 175, 1972–1988.e16 (2018).
Li, X. et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 20, 769–777 (2014).
DiMarco, R. L. et al. Engineering of three-dimensional microenvironments to promote contractile behavior in primary intestinal organoids. Integr. Biol. 6, 127–142 (2014).
Velasco, S. et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527 (2019).
Quadrato, G. et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53 (2017).
Paşca, A. M. et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678 (2015).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Múnera, J. O. et al. Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell Stem Cell 21, 51–64.e6 (2017).
McCracken, K. W. et al. Wnt/β-catenin promotes gastric fundus specification in mice and humans. Nature 541, 182–187 (2017).
McCracken, K. W. et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404 (2014).
Trisno, S. L. et al. Esophageal organoids from human pluripotent stem cells delineate Sox2 functions during esophageal specification. Cell Stem Cell 23, 501–515.e7 (2018).
Takebe, T. et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484 (2013).
Wong, A. P. et al. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 30, 876–882 (2012).
Huang, S. X. L. et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 32, 84–91 (2014).
Dye, B. R. et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife 4, (2015).
Chen, Y. W. et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 19, 542–549 (2017).
Kurmann, A. A. et al. Regeneration of thyroid function by transplantation of differentiated pluripotent stem cells. Cell Stem Cell 17, 527–542 (2015).
Longmire, T. A. et al. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell 10, 398–411 (2012).
Morizane, R. et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200 (2015).
Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015).
Ozone, C. et al. Functional anterior pituitary generated in self-organizing culture of human embryonic stem cells. Nat. Commun. 7, 10351 (2016).
Suga, H. et al. Self-formation of functional adenohypophysis in three-dimensional culture. Nature 480, 57–62 (2011).
Koehler, K. R., Mikosz, A. M., Molosh, A. I., Patel, D. & Hashino, E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature 500, 217–221 (2013).
Koehler, K. R. et al. Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat. Biotechnol. 35, 583–589 (2017).
Liu, X. P., Koehler, K. R., Mikosz, A. M., Hashino, E. & Holt, J. R. Functional development of mechanosensitive hair cells in stem cell-derived organoids parallels native vestibular hair cells. Nat. Commun. 7, 11508 (2016).
Nakano, T. et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785 (2012).
Lee, J. et al. Hair follicle development in mouse pluripotent stem cell-derived skin organoids. Cell Reports 22, 242–254 (2018).
Wimmer, R. A. et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature 565, 505–510 (2019).
Watson, C. L. et al. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 20, 1310–1314 (2014).
Phipson, B. et al. Evaluation of variability in human kidney organoids. Nat. Methods 16, 79–87 (2019).
Wu, H. et al. Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell 23, 869–881.e8 (2018).
van den Berg, C. W. et al. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Reports 10, 751–765 (2018).
Dye, B. R. et al. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. eLife 5, e19732 (2016).
Clevers, H. Modeling development and disease with organoids. Cell 165, 1586–1597 (2016).
McCauley, H. A. & Wells, J. M. Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development 144, 958–962 (2017).
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990).
Matano, M. et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 21, 256–262 (2015).
Drost, J. et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47 (2015).
IJspeert, J. E. G., Vermeulen, L., Meijer, G. A. & Dekker, E. Serrated neoplasia-role in colorectal carcinogenesis and clinical implications. Nat. Rev. Gastroenterol. Hepatol. 12, 401–409 (2015).
Fessler, E. et al. TGFβ signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol. Med. 8, 745–760 (2016).
Lannagan, T. R. M. et al. Genetic editing of colonic organoids provides a molecularly distinct and orthotopic preclinical model of serrated carcinogenesis. Gut 68, 684–692 (2019).
Kawasaki, K. et al. Chromosome engineering of human colon-derived organoids to develop a model of traditional serrated adenoma. Gastroenterology 158, 638–651 (2020).
Davis, H. et al. Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat. Med. 21, 62–70 (2015).
Drost, J. et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 358, 234–238 (2017).
Nadauld, L. D. et al. Metastatic tumor evolution and organoid modeling implicate TGFBR2 as a cancer driver in diffuse gastric cancer. Genome Biol. 15, 428 (2014).
van Es, J. H. & Clevers, H. Generation and analysis of mouse intestinal tumors and organoids harboring APC and K-Ras mutations. Methods Mol. Biol. 1267, 125–144 (2015).
Nanki, K. et al. Divergent routes toward Wnt and R-spondin niche independency during human gastric carcinogenesis. Cell 174, 856–869.e17 (2018).
Lee, J. et al. Reconstituting development of pancreatic intraepithelial neoplasia from primary human pancreas duct cells. Nat. Commun. 8, 14686 (2017).
Seino, T. et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell 22, 454–467.e6 (2018).
Liu, X. et al. Modeling Wnt signaling by CRISPR-Cas9 genome editing recapitulates neoplasia in human Barrett epithelial organoids. Cancer Lett. 436, 109–118 (2018).
Dekkers, J. F. et al. Modeling breast cancer using CRISPR-Cas9-mediated engineering of human breast organoids. J. Natl. Cancer Inst. 112, 540–544 (2020).
Artegiani, B. et al. Probing the tumor suppressor function of BAP1 in CRISPR-engineered human liver organoids. Cell Stem Cell 24, 927–943.e6 (2019).
Kim, J. et al. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Reports 3, 2088–2099 (2013).
Crespo, M. et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat. Med. 23, 878–884 (2017).
Huang, L. et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 21, 1364–1371 (2015).
Bian, S. et al. Genetically engineered cerebral organoids model brain tumor formation. Nat. Methods 15, 631–639 (2018).
Lee, D. F. et al. Modeling familial cancer with induced pluripotent stem cells. Cell 161, 240–254 (2015).
Han, T. et al. R-Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat. Commun. 8, 15945 (2017).
Zhang, H. et al. Gain-of-function RHOA mutations promote focal adhesion kinase activation and dependency in diffuse gastric cancer. Cancer Discov. 10, 288–305 (2019).
Bass, A. J. et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014).
Han, K. et al. CRISPR screens in cancer spheroids identify 3D growth-specific vulnerabilities. Nature 580, 136–141 (2020).
Lee-Six, H. et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature 574, 532–537 (2019).
Wang, Y. et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–160 (2014).
Blokzijl, F. et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016).
Roerink, S. F. et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 556, 457–462 (2018).
Behjati, S. et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature 513, 422–425 (2014).
Pollen, A. A. et al. Establishing cerebral organoids as models of human-specific brain evolution. Cell 176, 743–756.e17 (2019).
Christensen, S. et al. 5-Fluorouracil treatment induces characteristic T>G mutations in human cancer. Nat. Commun. 10, 4571 (2019).
Bolhaqueiro, A. C. F. et al. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat. Genet. 51, 824–834 (2019).
Kretzschmar, K. & Watt, F. M. Lineage tracing. Cell 148, 33–45 (2012).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Cortina, C. et al. A genome editing approach to study cancer stem cells in human tumors. EMBO Mol. Med. 9, 869–879 (2017).
Shimokawa, M. et al. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature 545, 187–192 (2017).
Barker, N. et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 (2009).
Schepers, A. G. et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337, 730–735 (2012).
Reya, T., Morrison, S. J., Clarke, M. F. & Weissman, I. L. Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001).
Sugimoto, S. et al. Reconstruction of the human colon epithelium in vivo. Cell Stem Cell 22, 171–176.e5 (2018).
de Sousa e Melo, F. et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 543, 676–680 (2017).
Fumagalli, A. et al. Plasticity of Lgr5-negative cancer cells drives metastasis in colorectal cancer. Cell Stem Cell 26, 569–578 (2020).
de Sousa E Melo, F. et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 9, 476–485 (2011).
Tian, H. et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259 (2011).
Yan, K. S. et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA 109, 466–471 (2012).
Heo, I. et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 3, 814–823 (2018).
Forbester, J. L. et al. Interaction of Salmonella enterica serovar Typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect. Immun. 83, 2926–2934 (2015).
Scanu, T. et al. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microbe 17, 763–774 (2015).
Wilson, S. S., Tocchi, A., Holly, M. K., Parks, W. C. & Smith, J. G. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol. 8, 352–361 (2015).
Zhang, Y. G., Wu, S., Xia, Y. & Sun, J. Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiol. Rep. 2, e12147 (2014).
Pleguezuelos-Manzano, C. et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580, 269–273 (2020).
Wroblewski, L. E. et al. Helicobacter pylori targets cancer-associated apical-junctional constituents in gastroids and gastric epithelial cells. Gut 64, 720–730 (2015).
Bertaux-Skeirik, N. et al. CD44 plays a functional role in Helicobacter pylori-induced epithelial cell proliferation. PLoS Pathog. 11, e1004663 (2015).
Finkbeiner, S. R. et al. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio 3, e00159–e12 (2012).
Ettayebi, K. et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393 (2016).
Hui, K. P. Y. et al. Tropism, replication competence, and innate immune responses of influenza virus: an analysis of human airway organoids and ex-vivo bronchus cultures. Lancet Respir. Med. 6, 846–854 (2018).
Wirbel, J. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 25, 679–689 (2019).
Byrne, A. T. et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer 17, 254–268 (2017).
Bleijs, M., van de Wetering, M., Clevers, H. & Drost, J. Xenograft and organoid model systems in cancer research. EMBO J. 38, e101654 (2019).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Weeber, F. et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc. Natl. Acad. Sci. USA 112, 13308–13311 (2015).
Gao, D. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014).
Jacob, F. et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell 180, 188–204 (2020).
Yan, H. H. N. et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell 23, 882–897.e11 (2018).
Schütte, M. et al. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat. Commun. 8, 14262 (2017).
Vlachogiannis, G. et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926 (2018).
Schumacher, D. et al. Heterogeneous pathway activation and drug response modelled in colorectal-tumor-derived 3D cultures. PLoS Genet. 15, e1008076 (2019).
Fujii, M. et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell 18, 827–838 (2016).
Ooft, S. N. et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 11, eaay2574 (2019).
Ganesh, K. et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat. Med. 25, 1607–1614 (2019).
Yao, Y. et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell 26, 17–26 (2020).
Steele, N. G. et al. An organoid-based preclinical model of human gastric cancer. Cell. Mol. Gastroenterol. Hepatol. 7, 161–184 (2019).
Seidlitz, T. et al. Human gastric cancer modelling using organoids. Gut 68, 207–217 (2019).
Tiriac, H. et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 8, 1112–1129 (2018).
Driehuis, E. et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl. Acad. Sci. USA 116, 26580–26590 (2019).
Mullenders, J. et al. Mouse and human urothelial cancer organoids: A tool for bladder cancer research. Proc. Natl. Acad. Sci. USA 116, 4567–4574 (2019).
Lee, S. H. et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 173, 515–528 (2018).
Puca, L. et al. Patient derived organoids to model rare prostate cancer phenotypes. Nat. Commun. 9, 2404 (2018).
de Witte, C. J. et al. Patient-derived ovarian cancer organoids mimic clinical response and exhibit heterogeneous inter- and intrapatient drug responses. Cell Reports 31, 107762 (2020).
Kijima, T. et al. Three-dimensional organoids reveal therapy resistance of esophageal and oropharyngeal squamous cell carcinoma cells. Cell. Mol. Gastroenterol. Hepatol. 7, 73–91 (2019).
Li, X. et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat. Commun. 9, 2983 (2018).
Driehuis, E. et al. Oral mucosal organoids as a potential platform for personalized cancer therapy. Cancer Discov. 9, 852–871 (2019).
Boretto, M. et al. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat. Cell Biol. 21, 1041–1051 (2019).
Li, L. et al. Human primary liver cancer organoids reveal intratumor and interpatient drug response heterogeneity. JCI Insight 4, 121490 (2019).
Nuciforo, S. et al. Organoid models of human liver cancers derived from tumor needle biopsies. Cell Reports 24, 1363–1376 (2018).
Broutier, L. et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 23, 1424–1435 (2017).
Calandrini, C. et al. An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nat. Commun. 11, 1310 (2020).
Kim, M. et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 10, 3991 (2019).
HCMI. Human Cancer Models Initiative. https://ocg.cancer.gov/programs/HCMI (2020).
De Roock, W. et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 11, 753–762 (2010).
Sonnenblick, A., de Azambuja, E., Azim, H. A. Jr. & Piccart, M. An update on PARP inhibitors—moving to the adjuvant setting. Nat. Rev. Clin. Oncol. 12, 27–41 (2015).
Vitale, I., Manic, G., Coussens, L. M., Kroemer, G. & Galluzzi, L. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 30, 36–50 (2019).
Joyce, J. A. & Fearon, D. T. T cell exclusion, immune privilege, and the tumor microenvironment. Science 348, 74–80 (2015).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437 (2013).
Kalbasi, A. & Ribas, A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 20, 25–39 (2020).
Wei, S. C., Duffy, C. R. & Allison, J. P. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 8, 1069–1086 (2018).
Biffi, G. et al. Il1-induced Jak/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. (2019).
Öhlund, D. et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214, 579–596 (2017).
Schnalzger, T. E. et al. 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 38, e100928 (2019).
Dijkstra, K. K. et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell 174, 1586–1598 (2018).
Pauli, C. et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 7, 462–477 (2017).
Weeber, F., Ooft, S. N., Dijkstra, K. K. & Voest, E. E. Tumor organoids as a pre-clinical cancer model for drug discovery. Cell Chem. Biol. 24, 1092–1100 (2017).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Garnett, M. J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575 (2012).
Edmondson, R., Broglie, J. J., Adcock, A. F. & Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 12, 207–218 (2014).
Jabs, J. et al. Screening drug effects in patient-derived cancer cells links organoid responses to genome alterations. Mol. Syst. Biol. 13, 955 (2017).
Verissimo, C. S. et al. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. eLife 5, e18489 (2016).
Skardal, A. et al. Drug compound screening in single and integrated multi-organoid body-on-a-chip systems. Biofabrication 12, 025017 (2020).
Geurts, M. H. et al. CRISPR-based adenine editors correct nonsense mutations in a cystic fibrosis organoid biobank. Cell Stem Cell 26, 503–510 (2020).
Shenoy, T. R. et al. CHD1 loss sensitizes prostate cancer to DNA damaging therapy by promoting error-prone double-strand break repair. Ann. Oncol. 28, 1495–1507 (2017).
Janda, C. Y. et al. Surrogate Wnt agonists that phenocopy canonical Wnt and β-catenin signalling. Nature 545, 234–237 (2017).
Mihara, E. et al. Active and water-soluble form of lipidated Wnt protein is maintained by a serum glycoprotein afamin/α-albumin. eLife 5, e11621 (2016).
Artegiani, B. et al. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat. Cell Biol. 22, 321–331 (2020).
Vaidyanathan, S. et al. High-efficiency, selection-free gene repair in airway stem cells from cystic fibrosis patients rescues CFTR function in differentiated epithelia. Cell Stem Cell 26, 161–171 (2020).
Michels, B. E. et al. Pooled in vitro and in vivo CRISPR-Cas9 screening identifies tumor suppressors in human colon organoids. Cell Stem Cell 26, 782–792 (2020).
Ringel, T. et al. Genome-scale CRISPR screening in human intestinal organoids identifies drivers of TGF-β resistance. Cell Stem Cell 26, 431–440 (2020).
Du, Y. et al. Development of a miniaturized 3D organoid culture platform for ultra-high throughput screening. J. Mol. Cell Biol. https://doi.org/10.1093/jmcb/mjaa036 (2020).
Acknowledgements
We thank members of the Kuo lab for discussions and Amanda Mah for figure artwork. This work was supported by US National Institutes of Health fellowship K00CA212433 (Y.-H. L.), a Swedish Research Council International Postdoctoral Fellowship (K. K.), and grants from the NIH (U01CA217851, U54CA224081, U01CA199241 and U19AI116484), Emerson Collective and Ludwig Cancer Research to C. J. K.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lo, YH., Karlsson, K. & Kuo, C.J. Applications of organoids for cancer biology and precision medicine. Nat Cancer 1, 761–773 (2020). https://doi.org/10.1038/s43018-020-0102-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-020-0102-y
This article is cited by
-
Establishment of tumor microenvironment-preserving organoid model from patients with intracranial meningioma
Cancer Cell International (2024)
-
Dual targeting of MAPK and PI3K pathways unlocks redifferentiation of Braf-mutated thyroid cancer organoids
Oncogene (2024)
-
Establishment and characterization of canine mammary tumoroids for translational research
BMC Biology (2023)
-
Tumor-derived systems as novel biomedical tools—turning the enemy into an ally
Biomaterials Research (2023)
-
Drug screening at single-organoid resolution via bioprinting and interferometry
Nature Communications (2023)