Abstract
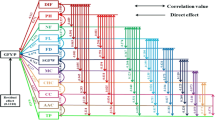
Capsicum L. is a genus of vegetables with a high global demand due to the pungency of its fruits. The species C. annuum L., C. frutescens L. and C. chinense Jacq. are the most cultivated and are closely related, belonging a group known as the annuum complex. Within these species, there are varieties with great morphological diversity that are widely exploited commercially. In this study, morphological measurements were performed on commercial chili pepper varieties, including Tabasco (C. frutescens), Cayenne (C. annuum) and Habanero (C. chinense), which are widely cultivated in the southwestern Colombian region, to generate a detailed phenotypic description and determine the correlation between traits. Additionally, microsatellite and SNP molecular markers were implemented to evaluate the genetic distances between them. The probability of forming hybrids between these varieties was also evaluated. Descriptive statistical parameters were estimated for the traits evaluated in forty plants of each commercial variety, and positive morphological correlations were found between the number of seeds, fruit mass and fruit length, as well as the independence or low correlation of this trait group versus the fruit diameter, day of first flower, number of fruits and productivity. The fruit capsaicin contents were estimated, with Habanero being the most pungent with 54.37 ± 5.83 mg/g. Molecular characterization using microsatellite markers and SNPs demonstrated the absence of heterozygous individuals and wide genetic distances between the commercial varieties evaluated; this outcome supported the impossibility of forming hybrids. The high genetic similarity among individuals within varieties could be useful to explore phenotypic plasticity in different environments. The methodology used here proved to be robust in testing trait correlation and cultivar genetic distancing, showing a look at the morphological and molecular relationships inside the genus which can be improved with the inclusion of more varieties. This information is especially useful for growers and breeders who wish to use and evaluate these plant materials.






Similar content being viewed by others
References
Andrews S (2018) FastQC v0.11.8: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc
Aguilar F, Aguilar B, García MA (2013) Análisis de medias generacionales para estimar parámetros genéticos de rendimiento en una cruza de pimentón y ají Cayenne (Capsicum annuum). Acta Agronómica 62(1):73–78
Avise JC (1994) Molecular markers natural history and evolution. Chapman and Hall, New York
Barbero GF, Liazid A, Palma M, Barroso CG (2008) Ultrasound-assisted extraction of capsaicinoids from peppers. Talanta 75:1332–1337
Barbosa GE, Carrizo-García C, Leiva-González S, Scaldaferro M, Reyes X (2019) Four new species of Capsicum (Solanaceae) from the tropical Andes and an update on the phylogeny of the genus. PLoS ONE 14(1):e0209792
Basu Ks, Krishna A (2003) Caspicum Medicinal and aromatic plants-Industrial profiles. Taylor & Francis, London (11 New Fetter Lane, London EC4P4EE. 277 pp)
Ben-Chaim A, Paran I (2000) Genetic analysis of quantitative traits in pepper (Capsicum annuum). J Amer Soc Hort Sci 125(1):66–70
Bosmali I, Ganopoulos I, Madesis P, Tsaftaris A (2012) Microsatellite and DNA-barcode regions typing combined with High Resolution Melting (HRM) analysis for food forensic uses: a case study on lentils (Lens culinaris). Food Res Int 46:141–147
Carrizo-García C, Sterpetti M, Volpi P, Ummarino M, Saccardo F (2013) Wild capsicums: identification and in situ analysis of Brazilian species. In: Lanteri S, Rotino GL (eds) Breakthroughs in the genetics and breeding of Capsicum and eggplant. Eucarpia, Turin, pp 205–213
Carrizo-García B, Sehr MHJ, Barbosa EM, Samuel GE, Moscone R, Ehrendorfer EA (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann Bot 118:35–51
Castañón-Nájera G, Latourneire-Moreno L, Lesher-Gordillo JM, Cruz-Lázaro E, Mendoza-Elos M (2010) Identificación de variables para caracterizar morfológicamente colectas de chile (Capsicum spp.) En Tabasco, México. Universidad y Ciencia Trópico Húmero 26(3):225–234
Castañón-Nájera G, Latourneire-Moreno L, Mendoza-Elos M, Vargas-López A, Cárdenas-Morales H (2008) Colección y caracterización de Chile (Capsicum spp.) en Tabasco. México Revista Internacional de Botánica Experimental 77:189–202
de Barboza GE, Bem Bianchetti L, Stehmann JR (2020) Capsicum carassense (Solanaceae), a new species from the Brazilian Atlantic Forest. PhytoKeys 140:125–138
Defaveri J, Viitaniemi H, Leder E, Merillä J (2013) Characterizing genic and nongenic molecular markers: comparison of microsatellites and SNPs. Mol Ecol Res 13:377–392
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresheaf tissue. Phytochem Bull 19:11–15
Ganopoulos I, Argiriou A, Tsaftaris A (2011) Microsatellite high resolution melting (SSR-HRM) analysis for authenticity testing of protected designation of origin (PDO) sweet cherry products. Food Control 22:532–541
González-Pérez S, Garcés-Claver A, Mallor C, de Sáenz Miera LE, Fayos O, Pomar F, Merino F, Silvar C (2014) New insights into Capsicum spp. relatedness and the diversification process of Capsicum annuum in Spain. PLoS ONE 9(12):e116276
Hamer K, Arrowsmith N, Gladis T (2003) Agrodiversity with emphasis on plant genetic resources. Naturwissenschaften 90:241–250
Hulse-Kemp A, Maheshwari S, Stoffel K, Hill TA, Jaffe D, Williams SR, Weisenfeld N, Ramakrishnan S, Kumar V, Shah P, Schatz MC, Church DM, Deynze AV (2018) Reference quality assembly of the 3.5-Gb genome of Capsicum annuum from a single linked-read library. Horticult Res 5:4
Iwai K, Suzuki T, Fujiwake H (1979) Formation and accumulation of pungent principle of hot pepper fruits, capsaicin and its analogues, in Capsicum annuum var annuum cv. Karayatsubusa at different stages of flowering. Agric Biol Chem 43:2496–2498
Jarret RL, Dang P (2004) Revisiting the waxy locus and the Capsicum annuum L. complex. Georgia J Sci 62(2):118–132
Jeong H-J, Jo YD, Park S-W, Kang B-C (2010) Identification of Capsicum species using SNP markers based on high resolution melting analysis. Genome 53:1029–1040
Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Justino EV, Fonseca MEN, Ferreira ME, Boiteux LS, Silva PP, Nascimento WM (2018) Estimate of natural cross-pollination rate of Capsicum annuum using a codominant molecular marker associated with fruit pungency. Genet Mol Res 17(1):gmr16039887
Kang JH, Yang HB, Jeong HS, Cheo P, Kwon JK, Kang BC (2014) Single nucleotide polymorphism marker discovery from transcriptome sequencing for marker-assisted backcrossing in Capsicum. Kor J Hortic Sci Technolo 32:535–543
Kumar S, Kumar R, Singh J (2006) Cayenne/American pepper (Capsicum species). In: Peter KV (ed) Handbook of herbs and spices. Woodhead, Cambridge, pp 299–312
Lee H-Y, Ro N-Y, Jeong H-J, Kwon J-K, Jo J, Ha Y, Jung A, Han J, Venkatesh J, Kang B-C (2016) Genetic diversity and population structure analysis to construct a core collection from a large Capsicum germplasm. BMC Genet 17:142
Lee JM, Nahm SH, Kim YM, Kim BD (2004) Characterization and molecular genetic mapping of microsatellite loci in pepper. Theoret Appl Genet 108:619–627
Marcos-Pérez D (2013) Caracterización molecular y análisis de diversidad genética en variedades de pimiento autóctonas de Galicia. Master´s Thesis. Universidad de Coruña. p 46
Martínez AC, Mejía MS, Ibarra DM, García MA, Cayón DG (2016) Response of Chili (Capsicum annuum L. Var. Cayena) to concentrations of N, P, K, Ca and Mg in Palmira, Valle del Cauca, Colombia. Revista Colombiana de Investigaciones Agroindustriales 3:40–48
Míngez-Mosquera MJ, Hornero-Mendes D (1994) Formation and transformation of pigments during the fruit ripening of Capsicum annuum cv. Bola and Agridulce. J Agric Food Chem 42:38–44
Nagy I, Stágel A, Sasvári Z, Röder M, Ganal M (2007) Development, characterization, and transferability to other Solanaceae of microsatellite markers in pepper (Capsicum annuum L.). Genome 50:668–688
Nwokem CO, Agbaji EB, Kagbu JA, Ekanem EJ (2010) Determination of Capsaicin content and pungency level of five different peppers grown in Nigeria. N Y Sci J 3(9):17–21
Olmstead RG, Bohs L, Migid HA, Santiago-Valentin E, Garcia VF, Collier SM (2008) A molecular phylogeny of Solanaceae. Taxon 57:1159–1181
Onus AN, Pickersgill B (2004) Unilateral incompatibility in Capsicum (Solanaceae): occurrence and taxonomic distribution. Annals Botany 94:289–295
Pardey C (2008) Caracterización y evaluación de Accesiones de Capsicum del banco de germoplasma de la Universidad Nacional sede Palmira y determinación del modo de herencia a Potyvirus. Doctoral thesis, Universidad Nacional.
Pardey C, García MA, Vallejo FA (2009) Evaluación agronómica de accesiones de Capsicum del banco de germoplasma de la Universidad Nacional de Colombia Sede Palmira. Acta Agronómica 58(1):23–28
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teacing and research. Mol Ecol Notes 6:288–295
Pérez-Castañeda LM, Castañón-Nájera G, Mayek-Pérez N (2008) Diversidad morfológica de chiles (Capsicum spp.) de Tabasco. México Cuadernos de Biodiversidad 27:11–22
Perkins B, Bushway R, Guthrie K, Fan T, Stewart B, Prince A, Williams M (2002) Determination of capsaicinoids in salsa by liquid chromatography and enzyme immunoassay. J AOAC Int 85(1):82–85
Pickersgill B (1969) The archeological record of chilli peppers (Capsicum spp) and the sequence of plant domestication in Peru. Am Antiq 34:53–61
Pickersgill B (1971) Relationships between weedy and cultivated forms in some species of chilli peppers (Genus Capsicum). Evolution 25:683–691
Pickersgill B, Heiser CB Jr, McNeill J (1979) Numerical taxonomic studies on variation and domestication in some species of Capsicum. In: Hawkes JG, Lester RN, Skelding AD (eds) The biology and taxonomy of the solanaceae. Academic Press, NY, pp 679–700
Pickergill B (1997) Genetic resources and breeding of Capsicum spp. Euphytica 96:129–133
Restrepo M, Llanos N, Fonseca CE (2007) Composición de las oleorresinas de dos variedades de ají picante (habanero y tabasco) obtenidas mediante lixiviación con solventes orgánicos. Revista Lasallista de Investigación 4(1):14–19
Rivera A, Monteagudo AB, Igartua E, Taboada A, García-Ulloa A, Pomar F, Riveiro-Leira M, Silvar C (2016) Assessing genetic and phenotypic diversity in pepper (Capsicum annuum L.) landraces from North-West Spain. Sci Hortic 203:1–11
Rodríguez E, Bolaños M, Menjivar JC (2010) Effect of the fertilization on the nutrition and yield of the red pepper (Capsicum spp.) in the Valley of the Cauca, Colombia. Acta Agronómica 59(1):55–64
Rodríguez-Maza MJ, Garcés-Claver AP, Kang S-W, Arnedo-Andrés B-C, M. S. (2012) A versatile PCR marker for pungency in Capsicum spp. Mol Breeding 30:889–898
Romero-Lozada M, Enciso CF, García SM, Wagner JJ, Puentes-Páramo YJ, Menjivar-Flores JC (2016) Efficiency of use of nutrients in hot pepper tabasco (Capsicum frutescens L.) and habanero (Capsicum chinense Jacq). Revista de Investigación Agraria y Ambiental 7(2):121–127
Sansaloni C, Petroli C, Jaccoud D, Carling J, Detering F, Grattapaglia D, Killian A (2011) Diversity Arrays Technology (DArT) and next-generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc 5(Suppl 7):P54
Särkinen T, Bohs L, Olmstead RG, Knapp S (2013) A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evol Biol 13:214
Savitha G, Slimath BP (1995) Effects of capsaicin on phospholipase A2 activity and superoxide generation in macrophages. Nutr Res 15:1417–1427
Snow AA, Whigham DF (1989) Cost of flower and fruit production in Tipularia discolor (Orchidaceae). Ecology 70(5):1286–1293
Stewart C (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14(5):748–750
Sun S, Frelich LE (2011) Flowering phenology and height growth pattern are associated with maximum plant height, relative growth rate and stem tissue mass density in herbaceous grassland species. J Ecol 99:991–1000
Tanksley SD (1984) High rates of cross-pollination in chile pepper. HortScience 19:580–582
Taranto F, D’Agostino N, Greco B, Cardi T, Tripodi P (2016) Genome-wide SNP discovery and population structure analysis in pepper (Capsicum annuum) using genotyping by sequencing. BMC Genomics 17:943
Tsykun T, Rellstab C, Dutech C, Sipos G, Prospero S (2017) Comparative assessment of SSR and SNP markers for inferring the population genetic structure of the common fungus Armillaria cepistipes. Heredity 119:371–380
Varshney RK, Graner A, Sorrells ME (2005) Genomics-assisted breeding for crop improvement. Trends Plant Sci 10:621–630
Walsh BM, Hoot SB (2001) Phylogenetic relationships of Capsicum (Solanaceae) using DNA sequences from two noncoding regions: the chloroplast atpB-rbcL spacer region and nuclear wasy introns. Int J Plant Sci 162:1409–1418
Willing E-M, Dreyer C, Oosterhout CV (2012) Estimates of genetic differentiation measured by FST do not necessarily require large sample sizes when using many SNP markers. PLoS ONE 8(7):e42649
Xanthopoulou A, Ganopoulos I, Koubouris G, Tsaftaris A, Sergendani C, Kalivas A, Madesis P (2014) Microsatellite high-resolution melting (SSR-HRM) analysis for genotyping and molecular characterization of an Olea europea germplasm collection. Plant Genetic Resources 12(3):273–277
Yumnam JS, Tyagi W, Pandey A, Meetei NT, Rai M (2012) Evaluation of genetic diversity ofchilli landraces from North Eastern India based on morphology, SSR markers and the Pun1 locus. Plant Mol Biol Rep 30(6):1470–1479
Zijlstra S, Purimahua C, Lindhout P (1991) Pollen tube growth in interspecific crosses between Capsicum species. HortScience 26(5):585–586
Acknowledgements
We thank the program Posgrado en Ciencias-Biología at the Universidad del Valle for their support during project implementation. We also thank the Escuela de Ingeniería Agrícola of the Universidad del Valle for allowing the development of the two phases of cultivation in 2016–2018 and its laboratory staff, specially to Hebert Hernández. We thank engineer Edilberto Muñoz and technician Ricardo Pereira from Hugo Restrepo and Co. for their support and help during project implementation. We thank the students José Enrique López Candelo, Héctor Cifuentes Silva and Juan Sebastián Reina Bolaños for their support during the field stage. To doctors Ana Belén Garcés Claver, Marisol Gordillo Suárez and Jaime Eduardo Muñoz Florez for their valuable comments. Special thanks to Alexander Ramírez for the support provided in the Molecular Biology Laboratory at Universidad del Valle. To professors José Hipólito Isaza Martínez and Alejandro Ortíz González of Departamento de Química from Universidad del Valle for their valuable support on capsaicin quantification. To Doctors Carolina Sansaloni, César Petroli, José Crossa and the professional Guadalupe Valdez from CIMMYT for their support in Illumina sequencing. Finally, we thank Luz Clemencia Aristizábal and Jenny Gallo for their support throughout the project. This work is dedicated to the memory of Carlos Alberto Duque Castaño, who always showed his admiration for the project.
Funding
This work was supported by the macro project: "Implementation of the Regional Center for Research and Innovation in Bioinformatics and Photonics, Cali, Valle del Cauca, Occidente" under Grant [BPIN 2013000100007].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest was reported by the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
South America, Colombia, Valle del Cauca, Cali. Geographic coordinates 3° 22′ 22.23″ N and 76° 31′ 47.82″ W.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Peñuela, M., Arias, L.L., Viáfara-Vega, R. et al. Morphological and molecular description of three commercial Capsicum varieties: a look at the correlation of traits and genetic distancing. Genet Resour Crop Evol 68, 261–277 (2021). https://doi.org/10.1007/s10722-020-00983-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-020-00983-8