Abstract
Human embryonic stem cells (hESCs) can differentiate to three germ layers within biochemical and biomechanical niches. The complicated mechanical environments in vivo could have diverse effects on the fate decision and biological functions of hESCs. To globally screen mechanosensitive molecules, three typical types of mechanical stimuli, i.e., tensile stretch, shear flow, and mechanical compression, were applied in respective parameter sets of loading pattern, amplitude, frequency, and/or duration, and then, iTRAQ proteomics test was used for identifying and quantifying differentially expressed proteins in hESCs. Bioinformatics analysis identified 37, 41, and 23 proteins under stretch pattern, frequency, and duration, 13, 18, and 41 proteins under shear pattern, amplitude, and duration, and 4, 0, and 183 proteins under compression amplitude, frequency, and duration, respectively, where distinct parameters yielded the differentially weighted preferences under each stimulus. Ten mechanosensitive proteins were commonly shared between two of three mechanical stimuli, together with numerous proteins identified under single stimulus. More importantly, functional GSEA and WGCNA analyses elaborated the variations of the screened proteins with loading parameters. Common functions in protein synthesis and modification were identified among three stimuli, and specific functions were observed in skin development under stretch alone. In conclusion, mechanomics analysis is indispensable to map actual mechanosensitive proteins under physiologically mimicking mechanical environment, and sheds light on understanding the core hub proteins in mechanobiology.








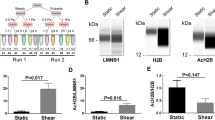
adopted from the literatures and re-plotted as the upper curves together with the lower tables. The dotted box between the curve and the table in each panel denotes the typical protein screened from the current mechanomics analysis. d Comparison of GSEA functional enrichment analysis from the current work with those typical functional tests in the literatures (Tannaz et al. 2014)
Similar content being viewed by others
Abbreviations
- ACY1:
-
Aminoacylase-1
- ANOVA:
-
Analysis of variance
- ANXA4:
-
Annexin A4
- BASP1:
-
Brain abundant membrane attached signal protein 1
- BP:
-
Biological process
- CC:
-
Cellular component
- CFL2:
-
Cofilin 2
- ES:
-
Enrichment score
- ESCs:
-
Embryonic stem cells
- EXOSC5:
-
Exosome component 5
- FC:
-
Fold change
- FDR:
-
False discovery rate
- GART:
-
Phosphoribosylglycinamide formyltransferase
- GO:
-
Gene ontology
- GSEA:
-
Gene set enrichment analysis
- hESCs:
-
Human embryonic stem cells
- HIST1H1B:
-
Histone cluster 1 H1 family member b
- HSP70:
-
Heat shock protein family A
- ITGB1:
-
Integrin beta1
- iTRAQ:
-
Isobaric tags for relative and absolute quantitation
- LAMB1/C1:
-
Lamin B1/C1
- MAPK:
-
Mitogen-activated kinase-like protein
- mESCs:
-
Mouse embryonic stem cells
- MDC1:
-
Mediator of DNA damage checkpoint 1
- MF:
-
Molecular function
- MFI:
-
Mean fluorescence intensity
- MTHFD1:
-
Methylenetetrahydrofolate dehydrogenase 1
- NES:
-
Normalized enrichment score
- PCA:
-
Principal component analysis
- PHD:
-
Plant homeodomain
- PSMs:
-
Protein spectrum matches
- Q-Q plot:
-
Quantile-sample quantile plot
- RhoA:
-
Ras homolog family member A
- RPL35A:
-
Ribosomal protein L35a
- RT:
-
Room temperature
- STON2:
-
Stonin 2
- TFI:
-
Total fluorescence intensity
- UHRF1:
-
Ubiquitin like with PHD and ring finger domains 1
- WGCNA:
-
Weighted gene co-expression network analysis
- YAP:
-
Yes associated protein 1
References
Adamo L, Naveiras O, Wenzel PL, McKinney-Freeman S, Mack PJ, Gracia-Sancho J, Suchy-Dicey A, Yoshimoto M, Lensch MW, Yoder MC, García-Cardeña G, Daley GQ (2009) Biomechanical forces promote embryonic haematopoiesis. Nature 459:1131–1135. https://doi.org/10.1038/nature08073
Behrndt M, Salbreux G, Campinho P, Hauschild R, Oswald F, Roensch J, Grill SW, Heisenberg CP (2012) Forces driving epithelial spreading in zebrafish gastrulation. Science 338:257–260. https://doi.org/10.1126/science.1224143
Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093. https://doi.org/10.1093/bioinformatics/btp101
Bonassar LJ, Grodzinsky AJ, Frank EH, Davila SG, Bhaktav NR, Trippel SB (2001) The effect of dynamic compression on the response of articular cartilage to insulin-like growth factor-I. J Orthop Res 19:11–17. https://doi.org/10.1016/s0736-0266(00)00004-8
Braet F, Shleper M, Paizi M, Brodsky S, Kopeiko N, Resnick N, Spira G (2004) Liver sinusoidal endothelial cell modulation upon resection and shear stress in vitro. Comp Hepatol 3:7–7. https://doi.org/10.1186/1476-5926-3-7
Chahine MN, Dibrov E, Blackwood DP, Pierce GN (2012) Oxidized LDL enhances stretch-induced smooth muscle cell proliferation through alterations in nuclear protein import. Can J Physiol Pharmacol 90:1559–1568. https://doi.org/10.1139/y2012-141
Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M (2001) AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451–464. https://doi.org/10.1016/s0092-8674(01)00578-5
Chin CH, Chen S-H, Wu HH, Ho CW, Ko MT, Lin CY (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8:S11. https://doi.org/10.1186/1752-0509-8-s4-s11
Chu TJ, Peters DG (2008) Serial analysis of the vascular endothelial transcriptome under static and shear stress conditions. Physiol Genomics 34:185–192. https://doi.org/10.1152/physiolgenomics.90201.2008
Cole RJ (1967) Cinemicrographic observations on the trophoblast and zona pellucida of the mouse blastocyst. J Embryol Exp Morphol 17:481–490
Conway A, Schaffer DV (2012) Biophysical regulation of stem cell behavior within the niche. Stem Cell Res Ther 3:50. https://doi.org/10.1186/scrt141
Cormier JT, Nieden NIZ, Rancourt DE, Kallos MS (2006) Expansion of undifferentiated murine embryonic stem cells as aggregates in suspension culture Bioreactors. Tissue Eng 12:3233–3245. https://doi.org/10.1089/ten.2006.12.3233
Costi M, Ferrari S (2001) Update on antifolate drugs targets. Curr Drug Targets 2:135–166. https://doi.org/10.2174/1389450013348669
Dado-Rosenfeld D, Tzchori I, Fine A, Chen-Konak L, Levenberg S (2015) Tensile forces applied on a cell-embedded three-dimensional scaffold can direct early differentiation of embryonic stem cells toward the mesoderm germ layer. Tissue Eng Part A 21:124–133. https://doi.org/10.1089/ten.tea.2014.0008
Dado D, Sagi M, Levenberg S, Zemel A (2012) Mechanical control of stem cell differentiation. Regen Med 7:101–116. https://doi.org/10.2217/rme.11.99
Derenzini M, Trere D, Pession A, Govoni M, Sirri V, Chieco P (2000) Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J Pathol 191:181–186. https://doi.org/10.1002/(SICI)1096-9896(200006)191:2<181:AID-PATH607>3.0.CO;2-V
Du V, Luciani N, Richard S, Mary G, Gay C, Mazuel F, Reffay M, Menasché P, Agbulut O, Wilhelm C (2017) A 3D magnetic tissue stretcher for remote mechanical control of embryonic stem cell differentiation. Nat Commun 8:400. https://doi.org/10.1038/s41467-017-00543-2
Eleuteri B, Aranda S, Ernfors P (2018) NoRC recruitment by H2A.X deposition at rRNA gene promoter limits embryonic stem cell proliferation. Cell Rep 23:1853–1866. https://doi.org/10.1016/j.celrep.2018.04.023
Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, Oria R, Kechagia JZ, Rico-Lastres P, Le Roux AL, Shanahan CM, Trepat XT, Navajas D, Garcia-Manyes S, Pere Roca-Cusachs P (2017) Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell 171:1397–1410. https://doi.org/10.1016/j.cell.2017.10.008
Fernandes AM, Fernandes TG, Diogo MM, da Silva CL, Henrique D, Cabral JMS (2007) Mouse embryonic stem cell expansion in a microcarrier-based stirred culture system. J Biotechnol 132:227–236. https://doi.org/10.1016/j.jbiotec.2007.05.031
Fok EYL, Zandstra PW (2005) Shear-controlled single-step mouse embryonic stem cell expansion and embryoid body-based differentiation. Stem Cells 23:1333–1342. https://doi.org/10.1634/stemcells.2005-0112
Gilmour D, Rembold M, Leptin M (2017) From morphogen to morphogenesis and back. Nature 541:311–320. https://doi.org/10.1038/nature21348
Heisenberg CP, Bellaiche Y (2013) Forces in tissue morphogenesis and patterning. Cell 153:948–962. https://doi.org/10.1016/j.cell.2013.05.008
Herranz R, Larkin OJ, Dijkstra CE, Hill RJA, Anthony P, Davey MR, Eaves L, van Loon JJWA, Medina FJ, Marco R (2012) Microgravity simulation by diamagnetic levitation: effects of a strong gradient magnetic field on the transcriptional profile of Drosophila melanogaster. BMC Genomics 13:52. https://doi.org/10.1186/1471-2164-13-52
Horiuchi R, Akimoto T, Hong Z, Ushida T (2012) Cyclic mechanical strain maintains Nanog expression through PI3K/Akt signaling in mouse embryonic stem cells. Exp Cell Res 318:1726–1732. https://doi.org/10.1016/j.yexcr.2012.05.021
Horner VL, Wolfner MF (2008) Mechanical stimulation by osmotic and hydrostatic pressure activates Drosophila oocytes in vitro in a calcium-dependent manner. Dev Biol 316:100–109. https://doi.org/10.1016/j.ydbio.2008.01.014
Huang HY, Nakayama Y, Qin K, Yamamoto K, Ando J, Yamashita J, Itoh H, Kanda K, Yaku H, Okamoto Y, Nemoto Y (2005) Differentiation from embryonic stem cells to vascular wall cells under in vitro pulsatile flow loading. J Artif Organs 8:110–118. https://doi.org/10.1007/s10047-005-0291-2
Hur YS, Park JH, Ryu EK, Yoon HJ, Yoon SH, Hur CY, Lee WD, Lim JH (2011) Effect of artificial shrinkage on clinical outcome in fresh blastocyst transfer cycles. Clin Exp Reprod Med 38:87–92. https://doi.org/10.5653/cerm.2011.38.2.87
Illi B, Scopece A, Nanni S, Farsetti A, Morgante L, Biglioli P, Capogrossi MC, Gaetano C (2005) Epigenetic histone modification and cardiovascular lineage programming in mouse embryonic stem cells exposed to laminar shear stress. Circ Res 96:501–508. https://doi.org/10.1161/01.RES.0000159181.06379.63
Ingber DE (2006) Cellular mechanotransduction: putting all the pieces together again. FASEB J 20:811–827. https://doi.org/10.1096/fj.05-5424rev
Ji JY, Jing H, Diamond SL (2003) Shear stress causes nuclear localization of endothelial glucocorticoid receptor and expression from the GRE promoter. Circ Res 92:279–285. https://doi.org/10.1161/01.res.0000057753.57106.0b
Jiang YK, Liu HW, Li H, Wang FJ, Cheng K, Zhou GD, Zhang WJ, Ye ML, Cao YL, Liu W, Zou HF (2011) A proteomic analysis of engineered tendon formation under dynamic mechanical loading in vitro. Biomaterials 32:4085–4095. https://doi.org/10.1016/j.biomaterials.2011.02.033
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. https://doi.org/10.1101/gr.092759.109
Kurpinski K, Chu J, Wang D, Li S (2009) Proteomic profiling of mesenchymal stem cell responses to mechanical strain and TGF-beta1. Cell Mol Bioeng 2:606–614. https://doi.org/10.1007/s12195-009-0090-6
Lam AP, Dean DA (2008) Cyclic stretch-induced nuclear localization of transcription factors results in increased nuclear targeting of plasmids in alveolar epithelial cells. J Gene Med 10:668–678. https://doi.org/10.1002/jgm.1187
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. https://doi.org/10.1186/1471-2105-9-559
Leeb C, Jurga M, McGuckin C, Forraz N, Thallinger C, Moriggl R, Kenner L (2011) New perspectives in stem cell research: beyond embryonic stem cells. Cell Prolif 44:9–14. https://doi.org/10.1111/j.1365-2184.2010.00725.x
Lemaitre C, Bickmore WA (2015) Chromatin at the nuclear periphery and the regulation of genome functions. Histochem Cell Biol 144:111–122. https://doi.org/10.1007/s00418-015-1346-y
Leung HW, Chen A, Choo ABH, Reuveny S, Oh SKW (2011) Agitation can induce differentiation of human pluripotent stem cells in microcarrier cultures. Tissue Eng Part C Methods 17:165–172. https://doi.org/10.1089/ten.tec.2010.0320
Li J, Zhang F, Chen JY (2011) An integrated proteomics analysis of bone tissues in response to mechanical stimulation. BMC Syst Biol 5(Suppl 3):S7. https://doi.org/10.1186/1752-0509-5-S3-S7
Li YSJ, Haga JH, Chien S (2005) Molecular basis of the effects of shear stress on vascular endothelial cells. J Biomech 38:1949–1971. https://doi.org/10.1016/j.jbiomech.2004.09.030
Liu Z, Wu HJ, Jiang KW, Wang YJ, Zhang WJ, Chu QQ, Li J, Huang HW, Cai T, Ji HB, Yang C, Tang N (2016) MAPK-mediated YAP activation controls mechanical-tension-induced pulmonary alveolar regeneration. Cell Rep 16:1810–1819. https://doi.org/10.1016/j.celrep.2016.07.020
Malone AMD, Batra NN, Shivaram G, Kwon RY, You L, Kim CH, Rodriguez J, Jair K, Jacobs CR (2007) The role of actin cytoskeleton in oscillatory fluid flow-induced signaling in MC3T3-E1 osteoblasts. Am J Physiol Cell Physiol 292:C1830–C1836. https://doi.org/10.1152/ajpcell.00352.2005
Mangala LS, Zhang Y, He Z, Emami K, Ramesh GT, Story M, Rohde LH, Wu H (2011) Effects of simulated microgravity on expression profile of microRNA in human lymphoblastoid cells. J Biol Chem 286:32483–32490. https://doi.org/10.1074/jbc.M111.267765
McKee C, Hong Y, Yao D, Chaudhry GR (2017) Compression induced chondrogenic differentiation of embryonic stem cells in three-dimensional polydimethylsiloxane scaffolds. Tissue Eng Part A 23:426–435. https://doi.org/10.1089/ten.tea.2016.0376
Mootha VK, Lindgren CM, Eriksson KF, Subramanian AS, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273. https://doi.org/10.1038/ng1180
Motosugi N, Bauer T, Polanski Z, Solter D, Hiiragi T (2005) Polarity of the mouse embryo is established at blastocyst and is not prepatterned. Genes Dev 19:1081–1092. https://doi.org/10.1101/gad.1304805
Mukherjee D, Gao M, O'Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J (2002) The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J 21:165–174. https://doi.org/10.1093/emboj/21.1.165
Nithiananthan S, Crawford A, Knock JC, Lambert DW, Whawell SA (2016) Physiological fluid flow moderates fibroblast responses to TGF-β1. J Cell Biochem 118:878–890. https://doi.org/10.1002/jcb.25767
Nsiah BA, Ahsan T, Griffiths S, Cooke M, Nerem RM, McDevitt TC (2014) Fluid shear stress pre-conditioning promotes endothelial morphogenesis of embryonic stem cells within embryoid bodies. Tissue Eng Part A 20:954–965. https://doi.org/10.1089/ten.tea.2013.0243
Patwari P, Lee RT (2008) Mechanical control of tissue morphogenesis. Circ Res 103:234–243. https://doi.org/10.1161/CIRCRESAHA.108.175331
Percharde M, Lin CJ, Yin Y, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen X, Ramalho-Santos M (2018) A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 174:391–405. https://doi.org/10.1016/j.cell.2018.05.043
Qi YX, Jiang J, Jiang XH, Wang XD, Ji SY, Han Y, Long DK, Shen BR, Yan ZQ, Chien S, Jiang ZL (2011) PDGF-BB and TGF-beta1 on cross-talk between endothelial and smooth muscle cells in vascular remodeling induced by low shear stress. Proc Natl Acad Sci USA 108:1908–1913. https://doi.org/10.1073/pnas.1019219108
Quinn TM, Grodzinsky AJ, Hunziker EB, Sandy JD (1998) Effects of injurious compression on matrix turnover around individual cells in calf articular cartilage explants. J Orthop Res 16:490–499. https://doi.org/10.1002/jor.1100160415
Reffay M, Parrini MC, Cochet-Escartin O, Ladoux B, Buguin A, Coscoy S, Amblard F, Camonis J, Silberzan P (2014) Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nat Cell Biol 16:217–223. https://doi.org/10.1038/ncb2917
Rogers RS, Dharsee M, Ackloo S, Sivak JM, Flanagan JG (2012) Proteomics analyses of human optic nerve head astrocytes following biomechanical strain. Mol Cell Proteomics. https://doi.org/10.1074/mcp.m111.012302
Saha S, Ji L, de Pablo JJ, Palecek SP (2008) TGFβ/Activin/Nodal pathway in inhibition of human embryonic stem cell differentiation by mechanical strain. Biophys J 94:4123–4133. https://doi.org/10.1529/biophysj.107.119891
Sanders MC, Way M, Sakai J, Matsudaira P (1996) Characterization of the actin cross-linking properties of the scruin-calmodulin complex from the acrosomal process of Limulus sperm. J Biol Chem 271:2651–2657. https://doi.org/10.1074/jbc.271.5.2651
Sargent CY, Berguig GY, Kinney MA, Hiatt LA, Carpenedo RL, Berson RE, McDevitt TC (2010) Hydrodynamic modulation of embryonic stem cell differentiation by rotary orbital suspension culture. Biotechnol Bioeng 105:611–626. https://doi.org/10.1002/bit.22578
Shafa M, Sjonnesen K, Yamashita A, Liu S, Michalak M, Kallos MS, Rancourt DE (2011) Expansion and long-term maintenance of induced pluripotent stem cells in stirred suspension bioreactors. J Tissue Eng Regen Med 6:462–472. https://doi.org/10.1002/term.450
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Shimko VF, Claycomb WC (2008) Effect of mechanical loading on three-dimensional cultures of embryonic stem cell-derived cardiomyocytes. Tissue Eng 14:49–58. https://doi.org/10.1089/ten.2007.0092
Shin JH, Tam BK, Brau RR, Lang MJ, Mahadevan L, Matsudaira P (2007) Force of an actin spring. Biophys J 92:3729–3733. https://doi.org/10.1529/biophysj.106.099994
Sorescu GP, Sykes M, Weiss D, Platt MO, Saha A, Hwang J, Boyd N, Boo YC, Vega JD, Taylor WR, Jo H (2003) Bone morphogenic protein 4 produced in endothelial cells by oscillatory shear stress stimulates an inflammatory response. J Biol Chem 278:31128–31135. https://doi.org/10.1074/jbc.m300703200
Stolberg S, McCloskey KE (2009) Can shear stress direct stem cell fate? Biotechnol Prog 25:10–19. https://doi.org/10.1002/btpr.124
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. https://doi.org/10.1073/pnas.0506580102
Tannaz NA, Ali SM, Nooshin H, Nasser A, Reza M, Amir A, Maryam J (2014) Comparing the effect of uniaxial cyclic mechanical stimulation and chemical factors on myogenin and Myh2 expression in mouse embryonic and bone marrow derived mesenchymal stem cells. Mol Cell Biomech 11:19–37
Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM (1998) Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147. https://doi.org/10.1126/science.282.5391.1145
Truong T, Shams H, Mofrad MRK (2015) Mechanisms of integrin and filamin binding and their interplay with talin during early focal adhesion formation. Integr Biol (Camb) 7:1285–1296. https://doi.org/10.1039/c5ib00133a
van Loon JJWA (2009) Mechanomics and physicomics in gravisensing Microgravity. Sci Technol 21:159–167
Vella D, Zoppis I, Mauri G, Mauri P, Di Silvestre D (2017) From protein-protein interactions to protein co-expression networks: a new perspective to evaluate large-scale proteomic data. EURASIP J Bioinform Syst Biol 2017:6. https://doi.org/10.1186/s13637-017-0059-z
Wang JW, Lü DY, Mao DB, Long M (2014) Mechanomics: an emerging field between biology and biomechanics. Protein Cell 5:518–531. https://doi.org/10.1007/s13238-014-0057-9
Wang JW, Wu Y, Zhang X, Zhang F, Lü DY, Shangguan B, Gao YX, Long M (2019) Flow-enhanced priming of hESCs through H2B acetylation and chromatin decondensation. Stem Cell Res Ther 10:349. https://doi.org/10.1186/s13287-019-1454-z
Ward DF Jr, Salasznyk RM, Klees RF, Backiel J, Agius P, Bennett K, Boskey A, Plopper GE (2007) Mechanical strain enhances extracellular matrix-induced gene focusing and promotes osteogenic differentiation of human mesenchymal stem cells through an extracellular-related kinase-dependent pathway. Stem Cells Dev 16:467–480. https://doi.org/10.1089/scd.2007.0034
Warren L, Buchanan JM (1957) Biosynthesis of the purines. XIX. 2-Amino-N-ribosylacetamide 5'-phosphate (glycinamide ribotide) transformylase. J Biol Chem 229:613–626
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer, New York
Wolfe RP, Ahsan T (2013) Shear stress during early embryonic stem cell differentiation promotes hematopoietic and endothelial phenotypes. Biotechnol Bioeng 110:1231–1242. https://doi.org/10.1002/bit.24782
Wolfe RP, Leleux J, Nerem RM, Ahsan T (2012) Effects of shear stress on germ lineage specification of embryonic stem cells. Integr Biol (Camb) 4:1263–1273. https://doi.org/10.1039/c2ib20040f
Xu J, Khor KA, Sui J, Zhang J, Tan TL, Chen WN (2008) Comparative proteomics profile of osteoblasts cultured on dissimilar hydroxyapatite biomaterials: an iTRAQ-coupled 2-D LC-MS/MS analysis. Proteomics 8:4249–4258. https://doi.org/10.1002/pmic.200800103
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. https://doi.org/10.1089/omi.2011.0118
Acknowledgements
The authors thank to Drs. Yan Zhang and Xiaohua Lei for technical assistance. LC-MS/MS analysis was conducted in Beijing Institute of Genomics, Chinese Academy of Sciences. FX-4000TM tension unit was used in Beijing Anzhen Hospital, Capital Medical University and FX-5000TM compression unit was used in Shandong Key Laboratory of Biophysics, Dezhou University. This work was supported by National Natural Science Foundation of China Grants 31661143044, 31627804, 31870931, 31470907, and Frontier Science Key Project of Chinese Science Academy grant QYZDJ-SSW-JSC018.
Author information
Authors and Affiliations
Contributions
ML, JW, FZ and DL conceived the project; FZ, JW and DL performed the experiments and analyzed the data; DL and LZ prepared the hESCs and analyzed the data; BS compiled the pump control software; YG designed the flow chamber; YW analyzed the data and prepared the figures; ML, ZF, JW, and DL wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10237_2020_1378_MOESM1_ESM.tif
Fig. S1. Integrality test of collected protein samples under distinct mechanical stimuli. Entire set of proteins used for iTRAQ analysis was defined using SDS-PAGE assay under tensile stretch (a-b), shear flow (c-d), or mechanical compression (e-f) with v (TIF 3026 kb)
10237_2020_1378_MOESM2_ESM.tif
Fig. S2. Reliability test of iTRAQ data sets under distinct mechanical stimuli. Plotted were linear regression analyses of PSMs (a, d, g), Unique Peptides (b, e, h), and Coverage (c, f, i) in intermittent-continuous stretch (a-c), pulsatile-steady shear (TIF 4794 kb)
10237_2020_1378_MOESM3_ESM.tif
Fig. S3. Normality test of fold change (FC) for all the identified proteins under distinct mechanical stimuli. Plotted were probability density functions (a-c) and Q-Q plots (d-f) under tensile stretch (a, d), shear flow (b, e), or mechanical compression (TIF 1734 kb)
10237_2020_1378_MOESM4_ESM.tif
Fig. S4. Circos plot for differential protein distribution on chromatins under tensile stretch (a), shear flow (b), or mechanical compression (c). Short black lines in the innermost circles denote genes encoding differential proteins (TIF 1293 kb)
10237_2020_1378_MOESM5_ESM.tif
Fig. S5. Enrichment analysis of GSEA presented in single mechanical stimuli. Sign (NES) in x-axis in B-D defines the direction of enrichment upon the definitions of FC values in (a). Plotted were the enriched terms for cellular component (CC) (b), biologi (TIF 3100 kb)
10237_2020_1378_MOESM6_ESM.tif
Fig. S6. GO analysis of correlated expression modules obtained from WGCNA analysis under distinct mechanical stimuli. (a-c) Enrichment of green module genes in CC (a), BP (b), and MF (c), respectively. (d-f) Enrichment of blue module genes in CC (d), BP (TIF 1941 kb)
10237_2020_1378_MOESM7_ESM.tif
Fig. S7. Ribosome-related GSEA enrichment under typical stretch (a-c), shear (d-f) and compression (g-l). Enriched at 0.1 Hz stretch were translational initiation (a), large ribosomal subunit (b) and ribosomal subunit (c), at 1.1 Pa shear were translation (TIF 6172 kb)
10237_2020_1378_MOESM8_ESM.tif
Fig. S8. Morphology analysis and DNA expression in nuclei under typical stretch, shear and compression. Cellular nuclei were stained by Hoechst 33342. Nuclear area (a, f, k), circularity (b, g, l), aspect ratio (c, h, m), and mean (d, i, n) or total (e, j (TIF 1229 kb)
Rights and permissions
About this article
Cite this article
Zhang, F., Wang, J., Lü, D. et al. Mechanomics analysis of hESCs under combined mechanical shear, stretch, and compression. Biomech Model Mechanobiol 20, 205–222 (2021). https://doi.org/10.1007/s10237-020-01378-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10237-020-01378-5