Abstract
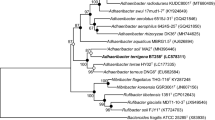
Strain MA2T was isolated from a soil sample from Gijang-gun, Busan in Korea. The strain, a Gram-stain-negative aerobic bacterium, is non-motile, ovoid- or rod-shaped, catalase- and oxidase-positive, and grows at NaCl concentrations 1% (w/v), at 15–30 °C (optimum 25 °C) and at pH 6–8.5 (optimum pH 7.5). The 16S rRNA gene sequence indicates that it belongs to the genus Adhaeribacter in the family Hymenobacteraceae. Phylogenetically, its closest relatives are Adhaeribacter terrae HY02T and Adhaeribacter terreus DNG6T, to which the strain shows 16S rRNA gene sequence similarity values of 96.6 and 96.0%, respectively. The major fatty acids (> 5% of the total fatty acids) of strain MA2T are C15:0 iso, C15:0 iso-G and summed feature 4 (anteiso-C17:1 B and/or iso-C17:1 I). The only detected isoprenoid quinone of strain MA2T is MK-7. The major polar lipid was phosphatidylethanolamine. The draft genome sequence of strain MA2T has a size of 4.9 Mkb. The genomic DNA G + C content was 46.9 mol%. Based on the phylogenetic, genotypic, phenotypic and chemotaxonomic data, the strain represents a novel species of the genus Adhaeribacter, for which the name Adhaeribacter soli sp. nov. is proposed. Strain MA2T (= KCTC 72630T = NBRC 114192T) is the type strain.

Similar content being viewed by others
References
Cappuccino JG, Sherman N (2002) Microbiology—a laboratory manual, 6th edn. Pearson Education, Inc., Benjamin Cummings, California
Chin CS, Alexander DH, Marks P et al (2013) Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569
Elderiny N, Lee J-J, Lee Y-H, Park S-J, Lee S-Y et al (2017) Adhaeribacter terrae sp. nov., a novel bacterium isolated from soil. Int J Syst Evol Microbiol 67:2922–2927
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by high performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469
Jeon Y-S, Kihyun L, Park S-C et al (2013) EzEditor: a versatile sequence alignment editor for both rRNA- and protein-coding genes. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijs.0.059360-0
Kim MK, Im W-T, Ohta H, Lee M, Lee S-T (2005) Sphingopyxis granuli sp. nov., a β-glucosidase-producing bacterium in the family Sphingomonadaceae in a-4 subclass of the Proteobacteria. J Microbiol 43:152–157
Kim MC, Kim CM, Kang OC, Zhang Y et al (2017) Hymenobacter rutilus sp. nov., isolated from marine sediment in the Arctic. Int J Syst Evol Microbiol 267:856–861
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Lee I, Kim YO, Park SC, Chun J (2015) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:1–14
Minnikin DE, O’Donnell AG, Goodfellow M et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Rickard AH, Stead AT, O’May GA et al (2005) Adhaeribacter aquaticus gen. nov., sp. nov., a gram-negative isolate from a potable water biofilm. Int J Syst Evol Microbiol 55:821–829
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Bio Evol 4:406–425
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol 44:846–849
Tamura K, Stecher G, Kumar S (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Tatusova T, DiCuccio M, Badretdin A et al (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44(14):6614–6624
Thompson JD, Gibson TJ, Plewniak F et al (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Weisburg WG, Barns SM, Pellerier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Acknowledgements
This work was supported by a research grant from Seoul Women’s University (2020) and by a grant from the National Institute of Fisheries Science (R2020031).
Author information
Authors and Affiliations
Contributions
All authors equally contributed to this work.
Corresponding authors
Ethics declarations
Conflict of interest
All authors certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Han, J.H., Kang, S. & Kim, M.K. Adhaeribacter soli sp. nov., a bacterium isolated from soil in Korea. Arch Microbiol 203, 163–168 (2021). https://doi.org/10.1007/s00203-020-01949-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-020-01949-1