Abstract
Type 2 cytokine responses promote parasitic immunity and initiate tissue repair; however, they can also result in immunopathologies when not properly restricted. Although basophilia is recognized as a common feature of type 2 inflammation, the roles basophils play in regulating these responses are unknown. Here, we demonstrate that helminth-induced group 2 innate lymphoid cell (ILC2) responses are exaggerated in the absence of basophils, resulting in increased inflammation and diminished lung function. Additionally, we show that ILC2s from basophil-depleted mice express reduced amounts of the receptor for the neuropeptide neuromedin B (NMB). Critically, NMB stimulation inhibited ILC2 responses from control but not basophil-depleted mice, and basophils were sufficient to directly enhance NMB receptor expression on ILC2s. These studies suggest that basophils prime ILC2s to respond to neuron-derived signals necessary to maintain tissue integrity. Further, these data provide mechanistic insight into the functions of basophils and identify NMB as a potent inhibitor of type 2 inflammation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Bulk RNA-seq and single-cell RNA-seq data are deposited in the Gene Expression Omnibus under accession code GSE150793. Source data are provided with this paper.
References
Schistosomiasis and soil-transmitted helminthiases: number of people treated in 2015. Wkly. Epidemiol. Rec. 91, 585–595 (2016).
Jourdan, P. M., Lamberton, P., Fenwick, A. & Addiss, D. G. Soil-transmitted helminth infections. Lancet 391, 252–265 (2018).
Jia, T.-W., Melville, S., Utzinger, J., King, C. H. & Zhou, X.-N. Soil-transmitted helminth reinfection after drug treatment: a systematic review and meta-analysis. PLoS Negl. Trop. Dis. 6, e1621 (2012).
Zhan, B. et al. Advancing a multivalent ‘pan-anthelmintic’ vaccine against soil-transmitted nematode infections. Expert Rev. Vaccines 13, 321–331 (2014).
Allen, J. E. & Maizels, R. M. Diversity and dialogue in immunity to helminths. Nat. Rev. Immunol. 11, 375–388 (2011).
Harris, N. L. & Loke, P. Recent advances in type-2-cell-mediated immunity: insights from helminth infection. Immunity 47, 1024–1036 (2017).
Gause, W. C., Wynn, T. A. & Allen, J. E. Type 2 immunity and wound healing: evolutionary refinement of adaptive immunity by helminths. Nat. Rev. Immunol. 13, 607–614 (2013).
Gieseck, R. L. III., Wilson, M. S. & Wynn, T. A. Type 2 immunity in tissue repair and fibrosis. Nat. Rev. Immunol. 18, 62–76 (2018).
Marsland, B. J., Kurrer, M., Reissmann, R., Harris, N. L. & Kopf, M. Nippostrongylus brasiliensis infection leads to the development of emphysema associated with the induction of alternatively activated macrophages. Eur. J. Immunol. 38, 479–488 (2008).
Sullivan, B. M. & Locksley, R. M. Basophils: a nonredundant contributor to host immunity. Immunity 30, 12–20 (2009).
Voehringer, D. Protective and pathological roles of mast cells and basophils. Nat. Rev. Immunol. 13, 362–375 (2013).
Kim, B. S. et al. Basophils promote innate lymphoid cell responses in inflamed skin. J. Immunol. 193, 3717–3725 (2014).
Siracusa, M. C., Kim, B. S., Spergel, J. M. & Artis, D. Basophils and allergic inflammation. J. Allergy Clin. Immunol. 132, 789–801 (2013).
Motomura, Y. et al. Basophil-derived interleukin-4 controls the function of natural helper cells, a member of ILC2s, in lung inflammation. Immunity 40, 758–771 (2014).
de Kleer, I. M. et al. Perinatal activation of the interleukin-33 pathway promotes type 2 immunity in the developing lung. Immunity 45, 1285–1298 (2016).
Cohen, M. et al. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell 175, 1031–1044.e18 (2018).
Siracusa, M. C. et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature 477, 229–233 (2011).
Webb, L. M. et al. The Notch signaling pathway promotes basophil responses during helminth-induced type 2 inflammation. J. Exp. Med. 216, 1268–1279 (2019).
Giacomin, P. R. et al. Thymic stromal lymphopoietin-dependent basophils promote Th2 cytokine responses following intestinal helminth infection. J. Immunol. 189, 4371–4378 (2012).
Ohnmacht, C. et al. Basophils orchestrate chronic allergic dermatitis and protective immunity against helminths. Immunity 33, 364–374 (2010).
Sullivan, B. M. et al. Genetic analysis of basophil function in vivo. Nat. Immunol. 12, 527–535 (2011).
Inclan-Rico, J. M. & Siracusa, M. C. First responders: innate immunity to helminths. Trends Parasitol. 34, 861–880 (2018).
Klose, C. S. N. et al. The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature 549, 282–286 (2017).
Cardoso, V. et al. Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature 549, 277–281 (2017).
Wallrapp, A. et al. The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature 549, 351–356 (2017).
Wallrapp, A. et al. Calcitonin gene-related peptide negatively regulates alarmin-driven type 2 innate lymphoid cell responses. Immunity 51, 709–723.e6 (2019).
Nagashima, H. et al. Neuropeptide CGRP limits group 2 innate lymphoid cell responses and constrains type 2 inflammation. Immunity 51, 682–695.e6 (2019).
Ohki-Hamazaki, H. Neuromedin B. Prog. Neurobiol. 62, 297–312 (2000).
Gajjar, S. & Patel, B. M. Neuromedin: an insight into its types, receptors and therapeutic opportunities. Pharmacol. Rep. 69, 438–447 (2017).
Noti, M. et al. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis. Nat. Med. 19, 1005–1013 (2013).
Saenz, S. A., Taylor, B. C. & Artis, D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol. Rev. 226, 172–190 (2008).
Stegle, O., Teichmann, S. A. & Marioni, J. C. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 16, 133–145 (2015).
Joseph, C. et al. Deciphering hematopoietic stem cells in their niches: a critical appraisal of genetic models, lineage tracing, and imaging strategies. Cell Stem Cell 13, 520–533 (2013).
Sutherland, T. E. et al. Chitinase-like proteins promote IL-17-mediated neutrophilia in a tradeoff between nematode killing and host damage. Nat. Immunol. 15, 1116–1125 (2014).
Chen, F. et al. Neutrophils prime a long-lived effector macrophage phenotype that mediates accelerated helminth expulsion. Nat. Immunol. 15, 938–946 (2014).
Chen, F. et al. An essential role for TH2-type responses in limiting acute tissue damage during experimental helminth infection. Nat. Med. 18, 260–266 (2012).
Minutti, C. M. et al. A macrophage-pericyte axis directs tissue restoration via amphiregulin-induced transforming growth factor beta activation. Immunity 50, 645–654.e6 (2019).
Ugajin, T. et al. FcεRI, but not FcγR, signals induce prostaglandin D2 and E2 production from basophils. Am. J. Pathol. 179, 775–782 (2011).
Siracusa, M. C., Comeau, M. R. & Artis, D. New insights into basophil biology: initiators, regulators, and effectors of type 2 inflammation. Ann. N. Y. Acad. Sci. 1217, 166–177 (2011).
Monticelli, L. A. et al. Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat. Immunol. 17, 656–665 (2016).
Zhou, Y. et al. Prostaglandin E2 inhibits group 2 innate lymphoid cell activation and allergic airway inflammation through E-prostanoid 4-cyclic adenosine monophosphate signaling. Front. Immunol. 9, 501 (2018).
Shimokawa, C. et al. Mast cells are crucial for induction of group 2 innate lymphoid cells and clearance of helminth infections. Immunity 46, 863–874.e4 (2017).
Roth, R. L. & Levy, D. A. Nippostrongylus brasiliensis: peripheral leukocyte responses and correlation of basophils with blood histamine concentration during infection in rats. Exp. Parasitol. 50, 331–341 (1980).
Ogilvie, B. M., Hesketh, P. M. & Rose, M. E. Nippostrongylus brasiliensis: peripheral blood leucocyte response of rats, with special reference to basophils. Exp. Parasitol. 46, 20–30 (1978).
Loffredo, L. F. et al. Eosinophil accumulation in postnatal lung is specific to the primary septation phase of development. Sci. Rep. 10, 4425 (2020).
Walunas, T. L. et al. CTLA-4 can function as a negative regulator of T cell activation. Immunity 1, 405–413 (1994).
Chikuma, S. et al. PD-1-mediated suppression of IL-2 production induces CD8+ T cell anergy in vivo. J. Immunol. 182, 6682–6689 (2009).
Klose, C. S. & Artis, D. Neuronal regulation of innate lymphoid cells. Curr. Opin. Immunol. 56, 94–99 (2019).
Nussbaum, J. C. et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248 (2013).
Flamar, A.-L. et al. Interleukin-33 induces the enzyme tryptophan hydroxylase 1 to promote inflammatory group 2 innate lymphoid cell-mediated immunity. Immunity 52, 606–619.e6 (2020).
Han, H. et al. IL-33 promotes gastrointestinal allergy in a TSLP-independent manner. Mucosal Immunol. 11, 394–403 (2018).
Wada, T. et al. Selective ablation of basophils in mice reveals their nonredundant role in acquired immunity against ticks. J. Clin. Investig. 120, 2867–2875 (2010).
Bouchery, T. et al. The study of host immune responses elicited by the model murine hookworms Nippostrongylus brasiliensis and Heligmosomoides polygyrus. Curr. Protoc. Mouse Biol. 7, 236–286 (2017).
Ohmori, K. et al. IL-3 induces basophil expansion in vivo by directing granulocyte-monocyte progenitors to differentiate into basophil lineage-restricted progenitors in the bone marrow and by increasing the number of basophil/mast cell progenitors in the spleen. J. Immunol. 182, 2835–2841 (2009).
Anderson, K. G. et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9, 209–222 (2014).
Jungblut, M., Oeltze, K., Zehnter, I., Hasselmann, D. & Bosio, A. Standardized preparation of single-cell suspensions from mouse lung tissue using the gentleMACS Dissociator. J. Vis. Exp. 1266 (2009).
Ewels, P. A. et al. The nf-core framework for community-curated bioinformatics pipelines. Preprint at Nat. Biotechnol. 38, 276–278 (2020).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Acknowledgements
We thank members of the Center for Immunity and Inflammation for discussions and critical reading, the New Jersey Medical School (NJMS) Flow Cytometry and Immunology Core Laboratory for technical assistance and the NJMS Histology Core and the NJMS Genomics Research Lab for bioinformatics assistance. We thank D. Voehringer for kindly providing the Mcpt8Cre mice and D. Sant’Angelo for providing the OP9-DL1 cell line. This work was supported by the National Institutes of Health (grant nos. RO1 AI123224 and RO1 AI131634 to M.C.S., T32AI125185 to C.B.S. and 3R01AI131634-02W1 to C.M.H.). J.M.I.R. is supported by the Mexican Council of Science & Technology (CVU 536876).
Author information
Authors and Affiliations
Contributions
J.M.I.R., J.J.P., N.V.P., C.M.H., C.B.S., A.D.L. and M.C.S. designed and performed the research. A.M.B. contributed to experimental design and data analysis, conceptualization and manuscript editing. J.M.I.R. and M.C.S. analyzed the experimental data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
Mark C. Siracusa is the founder and president of Nemagen Discoveries.
Additional information
Peer review information Jamie D. K. Wilson was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
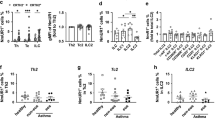
Extended Data Fig. 1 Basophils limit helminth-induced pulmonary inflammation.
a, Supernatant levels of IL-4, IL-5 and IL-13 from re-stimulated mesenteric lymph nodes (mLNs) isolated from control or basophil-depleted mice. Mucus production was evaluated in control and basophil-depleted mice on day 7 post-Nb infection by (b), periodic acid shiff (PAS) staining and (c), Muc5ac expression in the lungs by real-time PCR. d, Lung pathology was evaluated by H&E-stained sections with individual images digitally tiled together to provide a larger overview. P values were determined by two-tailed Student’s t-tests. a-d, Representative of at least 3 separate experiments with at least 5 mice per group. *P < 0.05, **P < 0.01, ***P < 0.001. (b), Illustrate data pooled from 2 separate experiments.
Extended Data Fig. 2 Basophil depletion results in elevated ILC2 responses.
a, Lung neutrophils and (b), eosinophils were quantified by flow cytometry on day 7 post-Nb infection in control or baso-dep mice. c, Representative flow cytometric gating strategy to evaluate neutrophils and eosinophils. d, ILC2s in the lung were quantified on day 7 post-Nb infection in control or baso-dep mice. Intracellular cytokine staining for (e, f), IL-5 and IL-13 was performed on lineage negative, CD90+, CD127+ ILC2s in lung on day 7 post-Nb infection and cytokine positive cells were quantified. g, Representative flow cytometric gating strategy to evaluate ILC2 populations. P values were determined by two-tailed Student’s t-tests. *P < 0.05, **P < 0.01, ***P < 0.001. a-g, Representative of at least 3 separate experiments with at least 5 mice per group.
Extended Data Fig. 3 Constitutive ablation of basophils is associated with increased ILC2 activation.
a, b, IL-5+ and IL-13+ ILC2s, as well as (c), eosinophils in the BAL and (d-f), lungs were quantified in control and Mcpt8Cre-4get mice that constitutively lack basophils, 7 days post-Nb infection. P values were determined by two-tailed Student’s t-tests. *P < 0.05, **P < 0.01, ***P < 0.001. a-f, Representative of at least 3 separate experiments with at least 5 mice per group.
Extended Data Fig. 4 Basophils are sufficient to limit helminth-induced ILC2 responses.
a, ILC2 numbers, (b), ILC2 production of IL-5, and (c), IL-13, as well as (d) eosinophil numbers were quantified in the lung on day 7 post-Nb infection in control mice, baso-dep mice, or baso-dep mice that received adoptive transfers of basophils. e, H&E staining of lung sections on day 7 post-Nb infection with individual images digitally tiled together to provide a larger overview. Mucus production in the lung was evaluated by (f), PAS staining of lung sections and (g), Muc5ac expression. P values were determined by two-tailed Student’s t-tests. *P < 0.05, **P < 0.01, ***P < 0.001. a-g, Representative of at least 3 separate experiments with at least 4 mice per group.
Extended Data Fig. 5 Basophils regulate ILC2s independently of adaptive lymphocytes.
Nb-infected Rag2-/- mice were treated with isotype control or the basophil-depleting antibody MAR-1 and (a), ILC2 responses and (b), eosinophilia were determined in the BAL and (c,d), lung on day 7 post-infection. (e), H&E staining of lung sections on day 7 post-Nb infection with individual images digitally tiled together to provide a larger overview. (f), Mucus production in the lung was evaluated by Muc5ac expression. P values were determined by two-tailed Student’s t-tests. *P < 0.05, **P < 0.01, ***P < 0.001. a-f, Representative of at least 3 separate experiments with at least 2 mice per naive groups and at least 4 mice per infected groups.
Extended Data Fig. 6 Elevated ILC2 responses are not associated with increased cytokine alarmin expression.
a-c, Expression of cytokine alarmins in the lungs of control and baso-dep mice was determined on day 7 post-Nb infection by real-time PCR. d, e, Numbers of IL-33-GFP+ type 1 and type 2 pneumocytes were evaluated in IL-33-GFP-reporter mice infected with Nb and treated with the basophil-depleting antibody MAR-1. Expression of (f), Il10 and (g), Areg in the lungs of control and baso-dep mice was determined on day 7 post-Nb infection by real-time PCR. Splenic basophils were sort-purified and cultured (O/N) with IL-3 and anti-IgE antibody and supernatant levels of (h), IL-6, (i), amphiregulin (Areg), and (j), IL-10 were evaluated by ELISA. P values were determined by two-tailed Student’s t-tests. *P < 0.05, **P < 0.01, ***P < 0.001. a-g, Representative of at least 2 separate experiments with at least 2 mice per naive groups and at least 5 mice per infected groups. h-j, Representative of at least 3 separate experiments with at least 5 individual samples of sort-purified basophils from 5 mice per experimental group.
Extended Data Fig. 7 Single cell RNAseq analysis of lung-resident ILC2s.
a, Uniform Manifold Approximation and Projection (UMAP) plot illustrating defined clusters of cells generated by single cell RNA-sequencing of lung-resident live ILC2 populations (CD45+Lin-CD90+CD127+) sort-purified from control (and basophil-depleted (baso-dep) mice 5 days post-Nb infection. b, Top 10 marker genes expressed by each cluster of ILC2s. c, Single-cell expression of Il5, Il13, Areg, Arg1, Il1rl1, and Il17rb in ILC cell clusters as defined in A. Horizontal bars represent mean normalized expression. P values were determined by Wilcoxon signed rank sum test. *P < 0.05, **P < 0.01, ***P < 0.001.
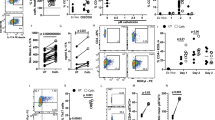
Extended Data Fig. 8 Analysis of NMBR expression in the hematopoietic compartment.
a, Heat map illustrating representative genes of interest expressed in control or baso-dep ILC2s. Surface NMBR expression by (b), CD4+ T cells, (c), alveolar macrophages, (d), non-alveolar macrophages, (e), neutrophils, and (f), eosinophils was determined in lung suspensions of naïve and mice infected with Nb 7 days prior. P values were determined by two-tailed Student’s t-tests. *P < 0.05, **P < 0.01, ***P < 0.001. (b-f), Representative of at least 3 separate experiments with at least 2 mice per naive groups and at least 4 mice per infected groups.
Extended Data Fig. 9 NMB-NMBR signaling suppresses helminth-induced ILC2 responses.
a, Schematic illustrating targeting strategy and placement of loxP cassettes upstream and downstream of exon 2 of the Nmbr gene. b-d, Type 2 cytokine expression in the lungs of NMBRfl/fl controls and NMBRfl/fl x Vav-iCre+ mice was determined on day 7 post-Nb infection by real-time PCR. e, IL-5+ and (f), IL-13+ ILC2s, as well as (g), eosinophils were quantified in the lungs of NMBRfl/fl x Vav-iCre+ mice 7 days post-Nb. Nb-infected Rag2-/- mice were treated with PBS or rNMB (i.t.) and (h, i), the percentage of IL-5+ and IL-13+ ILC2s were determined in the BAL and (j), the total number of IL-5+ and IL-13+ ILC2s were determined in the lung on day 7 post-infection. k, eosinophils and (l), neutrophils were determined in the lungs of Rag2-/- mice treated with PBS or rNMB on day 7-post infection. P values were determined by two-tailed Student’s t-tests. *P < 0.05, **P < 0.01, ***P < 0.001. b-l, Representative of 3 separate experiments with at least 3 mice per naive groups and at least 5 mice per infected groups.
Extended Data Fig. 10 Basophils are required for NMBR-mediated inhibition of ILC2s.
Sort-purified ILC2s were cultured (O/N) with vehicle or rNMB in the presence of IL-2 and IL-7 or IL-2, IL-7, and IL-33. a, b, The percentage of IL-5+ and IL-13+ ILC2s were quantified by intracellular staining. c, d, IL-5 and IL-13 levels in the supernatant were quantified by ELISA. Sort-purified ILC2s were cultured (O/N) alone or with activated basophils. e, Cytokine levels in the supernatant were monitored by ELISA and (f, g), cell proliferation was evaluated by CTV dilution 4 days post-culture. h, Heat map illustrating genes differentially expressed at 2.0-fold or higher between control or NMB-treated ILC2s. i, Heat map illustrating genes not differentially expressed in control or NMB-treated ILC2s. P values were determined by two-tailed Student’s t-tests. *P < 0.05, **P < 0.01, ***P < 0.001. a-g, Representative of at least 3 separate experiments with at least 5 individual samples of sort-purified ILC2s in each experimental group.
Supplementary information
Supplementary Table
Supplementary Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Inclan-Rico, J.M., Ponessa, J.J., Valero-Pacheco, N. et al. Basophils prime group 2 innate lymphoid cells for neuropeptide-mediated inhibition. Nat Immunol 21, 1181–1193 (2020). https://doi.org/10.1038/s41590-020-0753-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-0753-y
This article is cited by
-
The crosstalk between enteric nervous system and immune system in intestinal development, homeostasis and diseases
Science China Life Sciences (2024)
-
HTR2A agonists play a therapeutic role by restricting ILC2 activation in papain-induced lung inflammation
Cellular & Molecular Immunology (2023)
-
Single cell transcriptomics clarifies the basophil differentiation trajectory and identifies pre-basophils upstream of mature basophils
Nature Communications (2023)
-
Type 2 immunity in the brain and brain borders
Cellular & Molecular Immunology (2023)
-
Neuroimmunologie der allergischen Rhinitis Teil 2
HNO (2023)