Abstract
p53 is the most intensively studied tumour suppressor1. The regulation of p53 homeostasis is essential for its tumour-suppressive function2,3. Although p53 is regulated by an array of post-translational modifications, both during normal homeostasis and in stress-induced responses2,3,4, how p53 maintains its homeostasis remains unclear. UFMylation is a recently identified ubiquitin-like modification with essential biological functions5,6,7. Deficiency in this modification leads to embryonic lethality in mice and disease in humans8,9,10,11,12. Here, we report that p53 can be covalently modified by UFM1 and that this modification stabilizes p53 by antagonizing its ubiquitination and proteasome degradation. Mechanistically, UFL1, the UFM1 ligase6, competes with MDM2 to bind to p53 for its stabilization. Depletion of UFL1 or DDRGK1, the critical regulator of UFMylation6,13, decreases p53 stability and in turn promotes cell growth and tumour formation in vivo. Clinically, UFL1 and DDRGK1 expression are downregulated and positively correlated with levels of p53 in a high percentage of renal cell carcinomas. Our results identify UFMylation as a crucial post-translational modification for maintenance of p53 stability and tumour-suppressive function, and point to UFMylation as a promising therapeutic target in cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All of the source data supporting the findings of this study are available within this paper and/or from the corresponding author upon reasonable request. The human kidney renal clear cell carcinoma data were derived from The Cancer Genome Atlas Research Network (http://cancergenome.nih.gov/). Source data are provided with this paper.
References
Dolgin, E. The most popular genes in the human genome. Nature 551, 427–431 (2017).
Kastenhuber, E. R. & Lowe, S. W. Putting p53 in context. Cell 170, 1062–1078 (2017).
Vousden, K. H. & Prives, C. Blinded by the light: the growing complexity of p53. Cell 137, 413–431 (2009).
Kruse, J. P. & Gu, W. Modes of p53 regulation. Cell 137, 609–622 (2009).
Komatsu, M. et al. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 23, 1977–1986 (2004).
Tatsumi, K. et al. A novel type of E3 ligase for the Ufm1 conjugation system. J. Biol. Chem. 285, 5417–5427 (2010).
Daniel, J. & Liebau, E. The ufm1 cascade. Cells 3, 627–638 (2014).
Nahorski, M. S. et al. Biallelic UFM1 and UFC1 mutations expand the essential role of UFMylation in brain development. Brain 141, 1934–1945 (2018).
Tatsumi, K. et al. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat. Commun. 2, 181 (2011).
Zhang, M. et al. RCAD/Ufl1, a Ufm1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ. 22, 1922–1934 (2015).
Cai, Y. et al. UFBP1, a key component of the Ufm1 conjugation system, is essential for UFMylation-mediated regulation of erythroid development. PLoS Genet. 11, e1005643 (2015).
Egunsola, A. T. et al. Loss of DDRGK1 modulates SOX9 ubiquitination in spondyloepimetaphyseal dysplasia. J. Clin. Invest. 127, 1475–1484 (2017).
Yoo, H. M. et al. Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol. Cell 56, 261–274 (2014).
Gu, B. & Zhu, W. G. Surf the post-translational modification network of p53 regulation. Int. J. Biol. Sci. 8, 672–684 (2012).
Honda, R., Tanaka, H. & Yasuda, H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 (1997).
Scheffner, M., Huibregtse, J. M., Vierstra, R. D. & Howley, P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin–protein ligase in the ubiquitination of p53. Cell 75, 495–505 (1993).
Huang, Y. F., Wee, S., Gunaratne, J., Lane, D. P. & Bulavin, D. V. Isg15 controls p53 stability and functions. Cell Cycle 13, 2200–2210 (2014).
Weger, S., Hammer, E. & Heilbronn, R. Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 579, 5007–5012 (2005).
Maki, C. G., Huibregtse, J. M. & Howley, P. M. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 56, 2649–2654 (1996).
Lemaire, K. et al. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS ONE 6, e18517 (2011).
Liu, J. et al. A critical role of DDRGK1 in endoplasmic reticulum homoeostasis via regulation of IRE1α stability. Nat. Commun. 8, 14186 (2017).
Li, J. et al. Ufm1-specific ligase Ufl1 regulates endoplasmic reticulum homeostasis and protects against heart failure. Circ. Heart Fail. 11, e004917 (2018).
Wang, Z. et al. MRE11 UFMylation promotes ATM activation. Nucleic Acids Res. 47, 4124–4135 (2019).
Qin, B. et al. UFL1 promotes histone H4 UFMylation and ATM activation. Nat. Commun. 10, 1242 (2019).
Walczak, C. P. et al. Ribosomal protein RPL26 is the principal target of UFMylation. Proc. Natl Acad. Sci. USA 116, 1299–1308 (2019).
Wang, L. et al. UFMylation of RPL26 links translocation-associated quality control to endoplasmic reticulum protein homeostasis. Cell Res. 30, 5–20 (2020).
Nakamura, S., Roth, J. A. & Mukhopadhyay, T. Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol. Cell Biol. 20, 9391–9398 (2000).
Rodriguez, M. S., Desterro, J. M., Lain, S., Lane, D. P. & Hay, R. T. Multiple C-terminal lysine residues target p53 for ubiquitin–proteasome-mediated degradation. Mol. Cell Biol. 20, 8458–8467 (2000).
Kruse, J. P. & Gu, W. MSL2 promotes Mdm2-independent cytoplasmic localization of p53. J. Biol. Chem. 284, 3250–3263 (2009).
Chen, J., Marechal, V. & Levine, A. J. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell Biol. 13, 4107–4114 (1993).
Yuan, J., Luo, K., Zhang, L., Cheville, J. C. & Lou, Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 140, 384–396 (2010).
Noon, A. P. et al. p53 and MDM2 in renal cell carcinoma: biomarkers for disease progression and future therapeutic targets? Cancer 116, 780–790 (2010).
Riley, T., Sontag, E., Chen, P. & Levine, A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 (2008).
Hu, W., Feng, Z. & Levine, A. J. The regulation of multiple p53 stress responses is mediated through MDM2. Genes Cancer 3, 199–208 (2012).
Dai, C. & Gu, W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol. Med. 16, 528–536 (2010).
Beckerman, R. et al. Lysines in the tetramerization domain of p53 selectively modulate G1 arrest. Cell Cycle 15, 1425–1438 (2016).
Xu, J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr. Protoc. Mol. Biol. https://doi.org/10.1002/0471142727.mb2801s70 (2005).
Wu, J., Lei, G., Mei, M., Tang, Y. & Li, H. A novel C53/LZAP-interacting protein regulates stability of C53/LZAP and DDRGK domain-containing protein 1 (DDRGK1) and modulates NF-κB signaling. J. Biol. Chem. 285, 15126–15136 (2010).
Acknowledgements
We thank X. Li for providing the MDM2 and ubiquitin constructs, X. Xu for the UFM1 Lys-less construct and S. Bacchetti for critical reading of the manuscript. This work was supported by grants from the National Natural Science Foundation of China (31730020, 31871416, 81701380 and 31571409), Natural Science Foundation of Zhejiang Province of China (LY18C070001 and LY20H250002) and Hangzhou Science and Technology Bureau (20182014B01 and 20180533B27).
Author information
Authors and Affiliations
Contributions
J.L., D.G. and Y.-S.C. conceived of and designed the experiments. J.L., D.G., M.D., J.Y., H.W., L.S., L.X., J.B., C.L., Q.L. and J.Z. carried out the experiments. J.L., D.G., J.M., Q.Z., X.W., M.W. and Y.-S.C. analysed and interpreted the data. J.L., D.G. and Y.-S.C. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
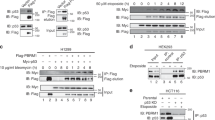
Extended Data Fig. 1 p53 interacts with UFL1 and DDRGK1.
a, Strategy for identification of binding proteins for UFL1 or DDRGK1. b, Proteins eluted from anti-Flag M2 affinity gel were subjected to SDS-PAGE followed by silver staining. c-e, Western blot analysis of the mutual interactions between p53, UFL1 and DDRGK1 in HEK293T cells by co-immunoprecipitation with anti-Flag M2 affinity gel. f, g, Western blot analysis of the mutual interactions between UFL1, DDRGK1 and p53 in HEK293T cells by co-immunoprecipitation with UFL1 (f) or DDRGK1 (g) antibody. Source data are available online.
Extended Data Fig. 2 The effects of UBA5, UFC1, UFL1, and DDRGK1 on p53 ufmylation in cells.
a, Western blot analysis of p53 ufmylation in HEK293T cells expressed the ufmylation system components in various combinations as indicated. b-e, Western blot analysis of p53 ufmylation in HEK293T cells with depletion of UFL1 (b), UBA5 (c), UFC1 (d), or DDRGK1 (e), respectively as indicated. Source data are available online.
Extended Data Fig. 3 p53 is ufmylated at Lys351, 357, 370 and 373 of C-terminal region.
a, The scheme diagram of p53-WT and its mutants. The Lys residues in p53 were replaced by Arg at the indicated positions. b, The ufmylation assay was performed to identity the ufmylation region of p53 in HEK293T cells expressed with p53-WT and its mutants as indicated in (a). c, Lys residues were replaced by Arg at indicated positions, and the ufmylation assay was performed in HEK293T cells. d, All Lys residues were replaced by Arg in p53 except one each of the five residues Lys351, 357, 370, 373 or 382 as indicated, and the ufmylation assay was performed in HEK293T cells. Source data are available online.
Extended Data Fig. 4 Ufmylation maintains p53 stability.
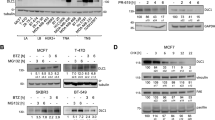
a, b, Q-PCR analysis of the relative mRNA expression of p53 and p21 in HeLa cells with UFL1 depletion (a), or DDRGK1 depletion (b). Error bars indicate mean ± s.e.m., n=3 for biological replicates. c, d, p53 stability was examined by western blot in HeLa cells with UFL1 (c), or DDRGK1 (d) depletion. The cells were treated with 100 µg ml−1 CHX for the indicated times, and the graph represents the quantification of the p53 protein levels. e, f, Western blot analysis of p53 expression in HCT116 (e), and HeLa (f) cells with UFL1 or DDRGK1 depletion in presence of MG132 (20 µM, 8 h). Source data are available online.
Extended Data Fig. 5 Ufmylation is required for p53 accumulation in DNA damage response.
a–d, Western blot analysis of p53 and p21 expression in HeLa cells with UFL1 or DDRGK1 depletion under DNA damage conditions (Doxo, 1 µM; Eto, 50µM) for indicated time. e, Western blot analysis of p53 and p21 expression in HCT116 p53+/+ and HCT116 p53−/− cells stably depletion of UFL1 or DDRGK1. f, Western blot analysis of p53 and p21 expression in HCT116 p53-/- cells stably expression of p53-WT or p53-4KR with UFL1 depletion. Source data are available online.
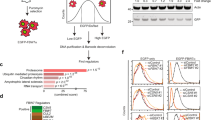
Extended Data Fig. 6 UFL1 and DDRGK1 are downregulated in renal cell carcinoma.
a-c, Immunohistochemical staining of UFL1 (a), DDRGK1 (b) and p53 (c) expression in forty pairs of RCC and adjacent normal tissues in tissue array. d, Percentages of the RCC specimens (a-c) showed low or high UFL1, DDRGK1 and p53 expression. (N, Normal tissue; C, cancer tissue). P values by a Chi-square (χ2) test, UFL1, P=2.16*10-14; DDRGK1, P=3.76*10-7; p53, P=8.29*10-16. Source data are available online.
Supplementary information
41556_2020_559_MOESM2_ESM.xlsx
Supplementary Tables Supplementary Table 1: Mass spectrometry information—UFL1 enrichment. The immunoprecipitates of Flag-UFL1 were resolved by SDS-PAGE and subjected to mass spectrometry for analysis. The spectra and unique peptide for each protein, protein name, protein description and molecular weight are listed. Supplementary Table 2: Mass spectrometry information—DDRGK1 enrichment. The immunoprecipitates of Flag-DDRGK1 were resolved by SDS-PAGE and subjected to mass spectrometry for analysis. The spectra and unique peptide for each protein, protein name, protein description and molecular weight are listed. Supplementary Table 3: Mass spectrometry information—p53 enrichment. The immunoprecipitates of Flag-p53 were resolved by SDS-PAGE and subjected to mass spectrometry for analysis. The spectra and unique peptide for each protein, protein name, protein description and molecular weight are listed. Supplementary Table 4: Antibody information. Information on the antibodies used in this study is provided in detail, including source, catalogue number, species, clone number (if available) and dilutions in distinct application. Supplementary Table 5: Plasmid information. Information on the plasmids used in the study is provided. Supplementary Table 6: Oligo information. Information on primers, including those used in quantitative PCR and siRNA interference, is provided in detail, including source and sequence data. Supplementary Table 7: Patient characteristics. Information on human research participants in the study is provided in detail, including patient ID, age, gender, p53 genotype and disease characteristics.
Source data
Source Data Fig. 1
Unprocessed western blots and gels.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots and gels.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
Liu, J., Guan, D., Dong, M. et al. UFMylation maintains tumour suppressor p53 stability by antagonizing its ubiquitination. Nat Cell Biol 22, 1056–1063 (2020). https://doi.org/10.1038/s41556-020-0559-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-020-0559-z
This article is cited by
-
Clinically relevant stratification of lung squamous carcinoma patients based on ubiquitinated proteasome genes for 3P medical approach
EPMA Journal (2024)
-
Ufmylation on UFBP1 alleviates non-alcoholic fatty liver disease by modulating hepatic endoplasmic reticulum stress
Cell Death & Disease (2023)
-
Programmed cell death 11 modulates but not entirely relies on p53-HDM2 loop to facilitate G2/M transition in colorectal cancer cells
Oncogenesis (2023)
-
Metabolic reprogramming and epigenetic modifications in cancer: from the impacts and mechanisms to the treatment potential
Experimental & Molecular Medicine (2023)
-
The ufmylation modification of ribosomal protein L10 in the development of pancreatic adenocarcinoma
Cell Death & Disease (2023)