Abstract
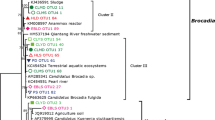
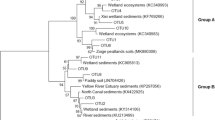
Upland soil clusters alpha and gamma (USCα and USCγ) are considered a major biological sink of atmospheric methane and are often detected in forest and grassland soils. These clusters are phylogenetically classified using the particulate methane monooxygenase gene pmoA because of the difficulty of cultivation. Recent studies have established a direct link of pmoA genes to 16S rRNA genes based on their isolated strain or draft genomes. However, whether the results of pmoA-based assays could be largely represented by 16S rRNA gene sequencing in upland soils remains unclear. In this study, we collected 20 forest soils across China and compared methane-oxidizing bacterial (MOB) communities by high-throughput sequencing of 16S rRNA and pmoA genes using different primer sets. The results showed that 16S rRNA gene sequencing and the semi-nested polymerase chain reaction (PCR) of the pmoA gene (A189/A682r nested with a mixture of mb661 and A650) consistently revealed the dominance of USCα (accounting for more than 50% of the total MOB) in 12 forest soils. A189f/A682r successfully amplified pmoA genes (mainly RA14 of USCα) in only three forest soils. A189f/mb661 could amplify USCα (mainly JR1) in several forest soils but showed a strong preferential amplification of Methylocystis and many other type I MOB groups. A189f/A650 almost exclusively amplified USCα (mainly JR1) and largely discriminated against Methylocystis and most of the other MOB groups. The semi-nested PCR approach weakened the bias of A189f/mb661 and A189f/A650 for JR1 and balanced the coverage of all USCα members. The canonical correspondence analysis indicated that soil NH4+-N and pH were the main environmental factors affecting the MOB community of Chinese forest soils. The RA14 of the USCα group prefers to live in soils with low pH, low temperature, low elevation, high precipitation, and rich in nitrogen. JR1’s preferences for temperature and elevation were opposite to RA14. Our study suggests that combining the deep sequencing of 16S rRNA and pmoA genes to characterize MOB in forest soils is the best choice.



Similar content being viewed by others
References
Angel R, Conrad R (2009) In situ measurement of methane fluxes and analysis of transcribed particulate methane monooxygenase in desert soils. Environ Microbiol 11(10):2598–2610
Aronson E, Allison S, Helliker BR (2013) Environmental impacts on the diversity of methane-cycling microbes and their resultant function. Front Microbiol 4:225
Baani M, Liesack W (2008) Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc Natl Acad Sci 105(29):10203–10208
Belova SE, Baani M, Suzina NE, Bodelier PLE, Liesack W, Dedysh SN (2011) Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ Microbiol Rep 3(1):36–46. https://doi.org/10.1111/j.1758-2229.2010.00180.x
Bodelier PL, Meima-Franke M, Hordijk CA, Steenbergh AK, Hefting MM, Bodrossy L, von Bergen M, Seifert J (2013) Microbial minorities modulate methane consumption through niche partitioning. ISME J 7(11):2214–2228. https://doi.org/10.1038/ismej.2013.99
Bourne DG, McDonald IR, Murrell JC (2001) Comparison of pmoA PCR primer sets as tools for investigating methanotroph diversity in three Danish soils. Appl Environ Microbiol 67(9):3802–3809
Cai Y, Zheng Y, Bodelier PLE, Conrad R, Jia Z (2016) Conventional methanotrophs are responsible for atmospheric methane oxidation in paddy soils. Nat Commun 7. https://doi.org/10.1038/ncomms11728
Costello AM, Lidstrom ME (1999) Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl Environ Microbiol 65(11):5066–5074
Danilova OV, Suzina NE, Van De Kamp J, Svenning MM, Bodrossy L, Dedysh SN (2016) A new cell morphotype among methane oxidizers: a spiral-shaped obligately microaerophilic methanotroph from northern low-oxygen environments. ISME J 10(11):2734–2743
Degelmann DM, Borken W, Drake HL, Kolb S (2010) Different atmospheric methane-oxidizing communities in European beech and Norway spruce soils. Appl Environ Microbiol 76(10):3228–3235. https://doi.org/10.1128/aem.02730-09
Deng Y, Che R, Wang F, Conrad R, Dumont M, Yun J, Wu Y, Hu A, Fang J, Xu Z, Cui X, Wang Y (2019) Upland soil cluster gamma dominates methanotrophic communities in upland grassland soils. Sci Total Environ 670:826–836. https://doi.org/10.1016/j.scitotenv.2019.03.299
Deng Y, Cui X, Lueke C, Dumont MG (2013) Aerobic methanotroph diversity in Riganqiao peatlands on the Qinghai-Tibetan Plateau. Environ Microbiol Rep 5(4):566–574. https://doi.org/10.1111/1758-2229.12046
Dumont MG, Lüke C, Deng Y, Frenzel P (2014) Classification of pmoA amplicon pyrosequences using BLAST and the lowest common ancestor method in MEGAN. Front Microbiol 5. https://doi.org/10.3389/fmicb.2014.00034
Dunfield PF, Belova SE, Vorob'ev AV, Cornish SL, Dedysh SN (2010) Methylocapsa aurea sp. nov., a facultative methanotroph possessing a particulate methane monooxygenase, and emended description of the genus Methylocapsa. Int J Syst Evol Microbiol 60(11):2659–2664
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Edwards CR, Onstott TC, Miller JM, Wiggins JB, Wang W, Lee CK, Cary SC, Pointing SB, Lau MCY (2017) Draft genome sequence of uncultured upland soil cluster <em>Gammaproteobacteria</em> gives molecular insights into high-affinity methanotrophy. Genome Announc 5(17):e00047–e00017. https://doi.org/10.1128/genomeA.00047-17
Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM et al (2010) Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464(7288):543–548
Fish JA, Chai BL, Wang Q, Sun YN, Brown CT, Tiedje JM et al (2013) FunGene: the functional gene pipeline and repository. Front Microbiol 4. https://doi.org/10.3389/fmicb.2013.00291
Frindte K, Maarastawi SA, Lipski A, Hamacher J, Knief C (2017) Characterization of the first rice paddy cluster I isolate, Methyloterricola oryzae gen. nov., sp. nov. and amended description of Methylomagnum ishizawai. Int J Syst Evol Microbiol
Henrysson T, McCarty PL (1993) Influence of the endogenous storage lipid poly-beta-hydroxybutyrate on the reducing power-availability during cometabolism of trichloroethylene and naphthalene by resting methanotrophic mixed cultures. Appl Environ Microbiol 59(5):1602–1606
Hirayama H, Abe M, Miyazaki M, Nunoura T, Furushima Y, Yamamoto H, Takai K (2014) Methylomarinovum caldicuralii gen. nov., sp. nov., a moderately thermophilic methanotroph isolated from a shallow submarine hydrothermal system, and proposal of the family Methylothermaceae fam. nov. Int J Syst Evol Microbiol 64(3):989–999
Holmes AJ, Costello A, Lidstrom ME, Murrell JC (1995) Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett 132(3):203–208
Holmes AJ, Roslev P, McDonald IR, Iversen N, Henriksen K, Murrell JC (1999) Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl Environ Microbiol 65(8):3312–3318
Horz HP, Rich V, Avrahami S, Bohannan BJM (2005) Methane-oxidizing bacteria in a California upland grassland soil: diversity and response to simulated global change. Appl Environ Microbiol 71(5):2642–2652. https://doi.org/10.1128/aem.71.5.2642-2652.2005
Judd CR, Koyama A, Simmons MP, Brewer P, von Fischer JC (2016) Co-variation in methanotroph community composition and activity in three temperate grassland soils. Soil Biol Biochem 95:78–86
Knief C (2015) Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol:6
Knief C, Dunfield PF (2005) Response and adaptation of different methanotrophic bacteria to low methane mixing ratios. Environ Microbiol 7(9):1307–1317
Knief C, Lipski A, Dunfield PF (2003) Diversity and activity of methanotrophic bacteria in different upland soils. Appl Environ Microbiol 69(11):6703–6714
Knief C, Vanitchung S, Harvey NW, Conrad R, Dunfield PF, Chidthaisong A (2005) Diversity of methanotrophic bacteria in tropical upland soils under different land uses. Appl Environ Microbiol 71(7):3826–3831
Kolb S (2009) The quest for atmospheric methane oxidizers in forest soils. Environ Microbiol Rep 1(5):336–346
Kolb S, Knief C, Dunfield PF, Conrad R (2005) Abundance and activity of uncultured methanotrophic bacteria involved in the consumption of atmospheric methane in two forest soils. Environ Microbiol 7(8):1150–1161
Kolb S, Knief C, Stubner S, Conrad R (2003) Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl Environ Microbiol 69(5):2423–2429
Kulichevskaya IS, Danilova OV, Tereshina VM, Kevbrin VV, Dedysh SN (2014) Descriptions of Roseiarcus fermentans gen. nov., sp. nov., a bacteriochlorophyll a-containing fermentative bacterium related phylogenetically to alphaproteobacterial methanotrophs, and of the family Roseiarcaceae fam. nov. Int J Syst Evol Microbiol 64(8):2558–2565
Lüke C, Frenzel P (2011) Potential of pmoA amplicon pyrosequencing for methanotroph diversity studies. Appl Environ Microbiol 77(17):6305–6309. https://doi.org/10.1128/aem.05355-11
Lüke C, Krause S, Cavigiolo S, Greppi D, Lupotto E, Frenzel P (2010) Biogeography of wetland rice methanotrophs. Environ Microbiol 12(4):862–872
Lau E, Ahmad A, Steudler PA, Cavanaugh CM (2007) Molecular characterization of methanotrophic communities in forest soils that consume atmospheric methane. FEMS Microbiol Ecol 60(3):490–500
Lau MCY, Stackhouse BT, Layton AC, Chauhan A, Vishnivetskaya TA, Chourey K, Ronholm J, Mykytczuk NCS, Bennett PC, Lamarche-Gagnon G, Burton N, Pollard WH, Omelon CR, Medvigy DM, Hettich RL, Pfiffner SM, Whyte LG, Onstott TC (2015) An active atmospheric methane sink in high Arctic mineral cryosols. ISME J 9:1880–1891. https://doi.org/10.1038/ismej.2015.13
McDonald IR, Bodrossy L, Chen Y, Murrell JC (2008) Molecular ecology techniques for the study of aerobic methanotrophs. Appl Environ Microbiol 74(5):1305–1315
Miroshnikov KK, Didriksen A, Naumoff DG, Huntemann M, Clum A, Pillay M et al (2017) Draft genome sequence of Methylocapsa palsarum NE2T, an obligate methanotroph from subarctic soil. Genome Announc 5(24):e00504–e00517
Nauer PA, Dam B, Liesack W, Zeyer J, Schroth MH (2012) Activity and diversity of methane-oxidizing bacteria in glacier forefields on siliceous and calcareous bedrock. Biogeosciences 9(6):2259–2274. https://doi.org/10.5194/bg-9-2259-2012
Nelson, D.W., and Sommers, L.E. (1996). “Total carbon, organic carbon, and organic matter,” in Methods of soil analysis, ed. D.L. Sparks. (Madison: Soil Science Society of America, American Society of Agronomy), 961-1010
Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ et al (2007) The vegan package. Commun Ecol Packag 10:631–637
Pieja AJ, Sundstrom ER, Criddle CS (2011) Poly-3-hydroxybutyrate metabolism in the type II methanotroph Methylocystis parvus OBBP. Appl Environ Microbiol 77(17):6012–6019. https://doi.org/10.1128/aem.00509-11
Pratscher J, Vollmers J, Wiegand S, Dumont MG, Kaster A-K (2018) Unravelling the identity, metabolic potential and global biogeography of the atmospheric methane-oxidizing upland soil cluster α. Environ Microbiol, n/a-n/a 20:1016–1029. https://doi.org/10.1111/1462-2920.14036
Qiu Q, Noll M, Abraham W-R, Lu Y, Conrad R (2008) Applying stable isotope probing of phospholipid fatty acids and rRNA in a Chinese rice field to study activity and composition of the methanotrophic bacterial communities in situ. ISME J 2(6):602–614
Radajewski S, Webster G, Reay DS, Morris SA, Ineson P, Nedwell DB, Prosser JI, Murrell JC (2002) Identification of active methylotroph populations in an acidic forest soil by stable-isotope probingc. Microbiology 148(8):2331–2342
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541
Shiau Y-J, Cai Y, Jia Z, Chen C-L, Chiu C-Y (2018) Phylogenetically distinct methanotrophs modulate methane oxidation in rice paddies across Taiwan. Soil Biol Biochem 124:59–69. https://doi.org/10.1016/j.soilbio.2018.05.025
Shrestha PM, Kammann C, Lenhart K, Dam B, Liesack W (2012) Linking activity, composition and seasonal dynamics of atmospheric methane oxidizers in a meadow soil. ISME J 6(6):1115–1126
Singh BK, Tate K (2007) Biochemical and molecular characterization of methanotrophs in soil from a pristine New Zealand beech forest. FEMS Microbiol Lett 275(1):89–97
Stoecker K, Bendinger B, Schöning B, Nielsen PH, Nielsen JL, Baranyi C, Toenshoff ER, Daims H, Wagner M (2006) Cohn's Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. PNAS 103(7):2363–2367
Stubner S (2002) Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen (TM) detection. J Microbiol Methods 50(2):155–164. https://doi.org/10.1016/s0167-7012(02)00024-6
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Theisen AR, Ali MH, Radajewski S, Dumont MG, Dunfield PF, McDonald IR, Dedysh SN, Miguez CB, Murrell JC (2005) Regulation of methane oxidation in the facultative methanotroph <i>Methylocella silvestris</i> BL2. Mol Microbiol 58(3):682–692
Tveit AT, Hestnes AG, Robinson SL, Schintlmeister A, Dedysh SN, Jehmlich N, von Bergen M, Herbold C, Wagner M, Richter A, Svenning MM (2019) Widespread soil bacterium that oxidizes atmospheric methane. Proc Natl Acad Sci:201817812. https://doi.org/10.1073/pnas.1817812116
Vekeman B, Kerckhof F-M, Cremers G, de Vos P, Vandamme P, Boon N, op den Camp HJM, Heylen K (2016) New Methyloceanibacter diversity from North Sea sediments includes methanotroph containing solely the soluble methane monooxygenase. Environ Microbiol 18(12):4523–4536. https://doi.org/10.1111/1462-2920.13485
Vigliotta G, Nutricati E, Carata E, Tredici SM, De Stefano M, Pontieri P et al (2007) Clonothrix fusca Roze 1896, a filamentous, sheathed, Methanotrophic γ-Proteobacterium. Appl Environ Microbiol 73(11):3556–3565. https://doi.org/10.1128/aem.02678-06
Vorobev AV, Baani M, Doronina NV, Brady AL, Liesack W, Dunfield PF, Dedysh SN (2011) Methyloferula stellata gen. nov., sp nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int J Syst Evol Microbiol 61:2456–2463. https://doi.org/10.1099/ijs.0.028118-0
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267
Wang Q, Quensen JF, Fish JA, Lee TK, Sun Y, Tiedje JM et al (2013) Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics using FrameBot, a new informatics tool. mBio 4(5)
Waring CL, Hankin SI, Griffith DWT, Kertesz MA, Kobylski V, Wilson NL, Coleman NV, Kettlewell G, Zlot R, Bosse M, Bell G (2017) Seasonal total methane depletion in limestone caves. Sci Rep 7(1):8314. https://doi.org/10.1038/s41598-017-07769-6
Zhang J-B, Cai Z-C, Zhu T-B, Yang W-Y, Müller C (2013) Mechanisms for the retention of inorganic N in acidic forest soils of southern China. Sci Rep 3:2342
Zhao R, Wang H, Cheng X, Yun Y, Qiu X (2018) Upland soil cluster γ dominates the methanotroph communities in the karst Heshang Cave. FEMS Microbiol Ecol 94(12). https://doi.org/10.1093/femsec/fiy192
Zheng Y, Huang R, Wang BZ, Bodelier PLE, Jia ZJ (2014) Competitive interactions between methane- and ammonia-oxidizing bacteria modulate carbon and nitrogen cycling in paddy soil. Biogeosciences 11(12):3353–3368
Zhou X-Q, Wang Y-F, Huang X-Z, Hao Y-B, Tian J-Q, Wang J-Z (2008) Effects of grazing by sheep on the structure of methane-oxidizing bacterial community of steppe soil. Soil Biol Biochem 40(1):258–261
Acknowledgments
We thank Prof. Jinbo Zhang at Nanjing Normal University for giving us these forest soil samples and also the basic information of these sampling sites. The four anonymous reviewers are gratefully acknowledged for constructive comments.
Funding
This work was supported by the National Science Foundation of China (41401294, 41877062, and 91751204), and Youth Innovation Promotion Association, CAS (2019311).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Cai, Y., Zhou, X., Shi, L. et al. Atmospheric Methane Oxidizers Are Dominated by Upland Soil Cluster Alpha in 20 Forest Soils of China. Microb Ecol 80, 859–871 (2020). https://doi.org/10.1007/s00248-020-01570-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01570-1