Abstract
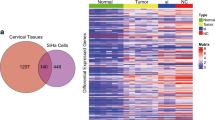
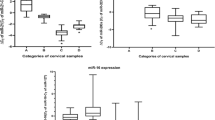
In this study we have investigated the effects of a tumour suppressor microRNA, miR-214, on gene expression in HPV-positive (CaSki) and HPV-negative cervical cancer cells (C33A) by RNA sequencing using next generation sequencing. The HPV-positive and HPV-negative cervical cancer cells were either miR-214-knocked-out or miR-214-overexpressed. Gene expression analysis showed that a total of 904 genes were upregulated and 365 genes were downregulated between HPV-positive and HPV-negative cervical cancer cells with a fold change of ±2. Furthermore, 11 differentially expressed and relevant genes (TNFAIP3, RAB25, MET, CYP1B1, NDRG1, CD24, LOXL2, CD44, PMS2, LATS1 and MDM1) which showed a fold change of ±5 were selected to confirm by real-time PCR. This study represents the first report of miR-214 on global gene expression in the context of HPV.





Similar content being viewed by others
Abbreviations
- 3ʹ-UTR:
-
3ʹ-untranslated region
- ATCC:
-
American Type Culture Collection
- CC:
-
cervical cancer
- CIN:
-
cervical intraepithelial neoplasia
- CRISPR:
-
clustered regularly interspaced short palindromic repeats
- DAVID:
-
the database for annotation, visualization and integrated discovery
- DEG:
-
differentially expressed genes
- DMEM:
-
Dulbecco’s Modified Eagle Medium
- FBS:
-
fetal bovine serum
- GO:
-
gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- miR:
-
microRNA
- NGS:
-
Next Generation sequencing
- NIAID:
-
National Institute of Allergy and Infectious Diseases
- qRT-PCR:
-
quantitative real-time PCR
- RIN:
-
RNA integrity number
- RNA-seq:
-
RNA-sequencing
- sgRNA:
-
single guide RNA
- SRA:
-
sequence read archive
References
Abba ML, Patil N, Leupold JH, Moniuszko M, Utikal J, Niklinski J and Allgayer H 2017 MicroRNAs as novel targets and tools in cancer therapy. Cancer Lett. 387 84–94
Agorastos T, Chatzistamatiou K, Katsamagkas T, Koliopoulos G, Daponte A, Constantinidis T, and Constantinidis TC 2015 Primary screening for cervical cancer based on high-risk human papillomavirus (HPV) detection and HPV 16 and HPV 18 genotyping, in comparison to cytology. PLoS ONE 10 e0119755
Berdasco M and Esteller M 2017 Crosstalk between non-coding RNAs and the epigenome in development. Chromatin Regul. Dyn. https://doi.org/10.1016/B978-0-12-803395-1.00009-5
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA and Jemal A 2018 Global cancer statistics GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 68 394–424
Byron SA, Keuren-Jensen KR Van, Engelthaler DM, Carpten JD and Craig DW 2016 Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat. Rev. Genet. 17 257–271
Chang I, Mitsui Y, Kim SK, et al. 2017 Cytochrome P450 1B1 inhibition suppresses tumorigenicity of prostate cancer via caspase-1 activation. Oncotarget 8 39087–39100
Deng J, Zhang W, Liu S, An H, Tan L and Ma L 2017 LATS1 suppresses proliferation and invasion of cervical cancer. Mol. Med. Rep. 15 1654–1660
Du B, Liu M, Li C, Geng X, Zhang X, Ning D and Liu M 2019 The potential role of TNFAIP3 in malignant transformation of gastric carcinoma. Pathol. Res. Pract. 215 152471
Frediani JN and Fabbri M 2016 Essential role of miRNAs in orchestrating the biology of the tumor microenvironment. Mol. Cancer 15 42
Ghittoni R, Accardi R, Hasan U, Gheit T, Sylla B, and Tommasino M 2010 The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes 40 1–13
Goodman A 2015 HPV testing as a screen for cervical cancer. BMJ 350 h2372
Hopman AHN and Ramaekers FCS 2017 Development of the uterine cervix and its implications for the pathogenesis of cervical cancer; In Pathology of the Cervix. Essentials of Diagnostic Gynecological Pathology (volume 3) (eds) Herrington C (Springer, Cham) pp 1–20
Imam N 2016 Computational analysis of human cancer related RNA-Seq data: A review. J. Appl. Comput. 1 30–37
Jaskowiak PA, Costa IG and Campello RJGB 2018 Clustering of RNA-Seq samples: Comparison study on cancer data. Methods 132 42–49
Jeong H, Lim KM, Kim KH, et al. 2019 Loss of Rab25 promotes the development of skin squamous cell carcinoma through the dysregulation of integrin trafficking. J. Pathol. 249 227–240
Jia J, Wang Z, Cai J and Zhang Y 2016 PMS2 expression in epithelial ovarian cancer is posttranslationally regulated by Akt and essential for platinum-induced apoptosis. Tumor Biol. 37 3059–3069
Kawai S, Fujii T, Kukimoto I, et al. 2018 Identification of miRNAs in cervical mucus as a novel diagnostic marker for cervical neoplasia. Sci. Rep. 8 7070
Kim J and Bang H 2016 Three common misuses of P-values. Dental Hypotheses 7 73
Li H, Zhang H, Lu G, Li Q, Gu J, Song Y, Gao S and Ding Y 2016 Mechanism analysis of colorectal cancer according to the microRNA expression profile. Oncol. Lett. 12 2329–2336
Mao Y, Shen J, Lu Y, et al. 2017 RNA sequencing analyses reveal novel differentially expressed genes and pathways in pancreatic cancer. Oncotarget 8 42537–42547
Mark D Van de, Kong D, Loncarek J and Stearns T 2015 MDM1 is a microtubule-binding protein that negatively regulates centriole duplication. Mol. Biol. Cell 26 3788–802
Mi L, Zhu F, Yang X, et al. 2017 The metastatic suppressor NDRG1 inhibits EMT, migration and invasion through interaction and promotion of caveolin-1 ubiquitylation in human colorectal cancer cells. Oncogene 36 4323–4335
Nagaya T, Nakamura Y, Okuyama S, Ogata F, Maruoka Y, Choyke PL, Allen C and Kobayashi H 2017 Syngeneic mouse models of oral cancer are effectively targeted by anti-CD44-based NIR-PIT. Mol. Cancer Res. 15 1667–1677
PEI Z, Zhu G, Huo X, et al. 2016 CD24 promotes the proliferation and inhibits the apoptosis of cervical cancer cells in vitro. Oncol. Rep. 35 1593–1601
Penna E, Orso F and Taverna D 2015 miR-214 as a key hub that controls cancer networks: small player, multiple functions. J. Invest. Dermatol. 135 960–9
Refaat T, Donnelly ED, Sachdev S, et al. 2017 c-Met overexpression in cervical cancer, a prognostic factor and a potential molecular therapeutic target. Am. J. Clin. Oncol. 40 590–597
Rupaimoole R and Slack FJ 2017 MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16 203–222
Salvador F, Martin A, López-Menéndez C, et al. 2017 Lysyl oxidase-like protein LOXL2 promotes lung metastasis of breast cancer. Cancer Res. 77 5846–5859
Santos JMO, Peixoto da Silva S, Costa NR, Gil da Costa RM and Medeiros R 2018 The role of MicroRNAs in the metastatic process of high-risk HPV-induced cancers. Cancers 10 493
Sen P, Ganguly P and Ganguly N 2018 Modulation of DNA methylation by human papillomavirus E6 and E7 oncoproteins in cervical cancer. Oncol. Lett. 15 11–22
Sochor M, Basova P, Pesta M, Dusilkova N, Bartos J, Burda P, Pospisil V and Stopka T 2014 Oncogenic MicroRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer 14 448
Tomasetti M, Amati M, Santarelli L and Neuzil J 2016 MicroRNA in metabolic re-programming and their role in tumorigenesis. Int. J. Mol. Sci. 17 754
Uhlen M, Zhang C, Lee S, et al. 2017 A pathology atlas of the human cancer transcriptome. Science 357 eaan2507
Acknowledgements
The funding from SERB, India, is duly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Saumitra Das.
Editorial Responsibility: Saumitra Das
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sen, P., Ganguly, P., Kulkarni, K.K. et al. Differential transcriptome analysis in HPV-positive and HPV-negative cervical cancer cells through CRISPR knockout of miR-214. J Biosci 45, 104 (2020). https://doi.org/10.1007/s12038-020-00075-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12038-020-00075-w