Abstract
Tuber species may be regarded as complex microhabitats hosting diverse microorganisms inside their fruiting bodies. Here, we investigated the structure of microbial communities inhabiting the gleba of wild growing (in stands) T. aestivum, using Illumina sequencing and culture-based methods. The two methods used in combination allowed to extract more information on complex microbiota of Tuber aestivum gleba. Analysis of the V3–V4 region of 16S rDNA identified nine phyla of bacteria present in the gleba of T. aestivum ascomata, mostly Proteobacteria from the family Bradyrhizobiaceae. Our results ideally match the earlier data for other Tuber species where the family Bradyrhizobiaceae was the most represented. The ITS1 region of fungal rDNA represented six alien fungal species belonging to three phyla. To complement the metagenomic analysis, cultivable fungi and bacteria were obtained from the gleba of the same T. aestivum fruiting bodies. The identified fungi mostly belong to the phylum Basidiomycota and same to Ascomycota. Analysis of cultivable bacteria revealed that all the specimens were colonized by different strains of Bacillus. Fungal community inhabiting T. aestivum fruiting bodies was never shown before.
Similar content being viewed by others
Introduction
Truffles are hypogeous ascomycetous fungi belonging to the genus Tuber, which form ectomycorrhizae with trees and shrubs. Some Tuber species, produce edible fruiting bodies with a unique flavor and texture and can be regarded as commercial. Two species, Tuber magnatum Pico, the white truffle, and Tuber melanosporum Vittad., the black truffle, are the most valued by gourmets for their organoleptic properties (Buzzini et al. 2005; Zambonelli et al. 2015). The occurrence of T. melanosporum Vittad. is noted mainly from Italy, France, Spain and Balkan countries, such as Croatia and Slovenia and the white truffle (T. magnatum Pico) grows exclusively in Italy, Croatia, Slovenia and Hungary (Mello et al. 2006; Büntgen et al. 2011; Pieroni 2016). Increasing attention is drawn toward other Tuber species, specifically Tuber aestivum Vittad., which exhibits a much wider distribution than any other truffle species. T. aestivum has been found in nearly all European countries and beyond, with habitats reaching as far as China and North Africa (Marocco) (Stobbe et al. 2013; Zambonelli et al. 2012). Recent overexploitation and disruption of the natural habitat of the truffle, deforestation, the loss of host plants within forests, the replacement of natural forests with plantations of species that are poor hosts, global warming, acid rain and the loss of expertise during two World Wars as to where and how to harvest mushrooms, particularly truffles (Wang et al. 2013) result in their decreased production. Hence efficient cultivation methods of the valued fungi are highly desirable. In this aspect, a detailed understanding of the interdependences between truffles and their environment is needed. At present, only T. melanosporum, T. aestivum and T. borchii Vittad. are collected from truffle plantations (Hall et al. 2003; Stobbe et al. 2013; Wang et al. 2013; Zambonelli et al. 2015). In contrast, production of T. magnatum in controlled conditions has failed so far (Mello et al. 2017; Zambonelli et al. 2015).
The fruiting bodies of the truffle are hypogeous, it is, therefore, highly likely that soil microorganisms affect their formation (Salerni et al. 2014). During their complex life cycle, truffles establish symbiotic interactions with bacteria (Archaea and Eubacteria) (Antony- Babu et al. 2014; Barbieri et al. 2000, 2005, 2007; Gryndler et al. 2013), fungi (yeasts and filamentous fungi) (Buzzini et al. 2005; Pacioni et al. 2007) and viruses (Stielow and Menzel 2010), at all stages of their development, which include (i) a symbiotic stage in association with the host plant (ectomycorrhiza), (ii) a sexual stage (fruiting bodies) and (iii) a “free living mycelial stage”.
The truffle fruiting body is built of mycelium, in which the outer cells differentiate into a protective layer (peridium) (Pacioni et al. 1995; Zarivi et al. 2015). In T. aestivum, the peridium is robust and composed of sclerified and melanized cells with pores forming an outlet of veins which represent authentic entryways (Pacioni 1990). To date, microbes and microbial communities from truffles have been characterized with culture-dependent and independent techniques (Antony-Babu et al. 2014; Barbieri et al. 2000, 2005, 2007, 2016; Benucci and Bonito 2016; Buzzini et al. 2005; Citterio et al. 1995; Gryndler et al. 2013; Nazzaro et al. 2007; Pacioni et al. 2007; Rivera et al. 2010; Sbrana et al. 2002; Splivallo et al. 2015, Stielow and Menzel 2010; Stielow et al. 2011a, b, 2012; Zacchi et al. 2003). Those studies focused mainly on bacteria and rarely on yeast and fungi inhabiting T. magnatum, T. melanosporum, and T. borchii. Pacioni et al. (2007) also investigated fungi from the fruiting body of T. aestivum. Despite those studies, the truffle-inhabiting fungi and bacteria are still poorly characterized, especially those associated with T. aestivum. A systematic investigation of fungi living inside the truffle fruiting body has never been performed. Only in recent years, studies on this topic have been initiated in relation to diseases and the shelf life of truffles (Pacioni and Leonardi 2016). Here, we present a microbiome analysis of the gleba of T. aestivum fruiting bodies harvested in southern Poland using culture-dependent and culture-independent methods.
Material and methods
Fruiting bodies of T. aestivum were harvested by manual digging from the natural habitats in September 2017, in two localities of Nida Basin (southern Poland). The fruiting bodies signed as 1–3 were sampled in the broadleaved forest with Quercus petraea (Matt.) Liebl., Acer pseudoplatanus L., Carpinus betulus L. at the altitude of 290–296 m a.s.l. and another three fruiting bodies (signed as 4–6) came from the thicket with Carpinus betulus L., Acer campestre L. and Populus tremula L. which grew at 227–228 m a.s.l. The sites were about 50 km apart. Average annual precipitation and temperature in the Nida Basin region (latitude 50°25´N and longitude 20°19´E) are as follow: average annual precipitation is 600 mm, and average temp. 8.0 °C. Soil conditions of the T. aestivum growing sites were characterized previously by Hilszczańska et al. (2008, 2014).
The harvested truffles were unwashed, separately packed into vacuum boxes and immediately transferred to the laboratory for analysis (Fig. 1). The species identity was determined by DNA analysis (Martin and Rygiewicz, 2005; Weden et al. 2005). DNA was extracted from gleba of T. aestivum using Plant DNA Mini Kit (Syngen Biotech, Poland). Extracted DNA was used as the template for amplification of the ITS region using universal primers ITS1 and ITS4 (White et al. 1990) and a standard PCR protocol (Martin and Rygiewicz 2005). Amplified products were separated by electrophoresis on an agarose gel, isolated from the gel by Qiaex II Gel extraction kit (Qiagen) and sequenced in the Institute of Biochemistry and Biophysics PAS using ABI3730XL DNA Analyzer (Thermo Fisher Scientific Inc. Waltham, MA, USA).
Tuber aestivum ascomata a, b and asci with ascospores c, d. A—T. aestivum fruiting bodies. b—fruiting bodies cut for gleba isolation. c—asci with ascospores stained with Atto488 dye conjugated with wheat germ lectin (Sigma-Aldrich) which binds to chitin in asci and ascospores. d—Evaluation of spores melanization in the studied specimens. Samples were examined using a Delta Optical microscope at 40 × magnification. Bar = 50 µm
The obtained sequences were compared to the sequences of Tuber aestivum isolated in Poland KX028767, KX028766 (Hilszczańska et al. 2016) and deposited in the Genbank database NCBI (Benson et al. 2013). For analysis only, fully mature ascocarps with 71–100% asci containing melanized spores (Büntgen et al. 2017) were chosen. Melanization was evaluated under a Delta Optical microscope (Fig. 1d).
Preparation of 16S rDNA and ITS1 amplicon libraries, DNA sequencing and data analysis
Six T. aestivum ascocarps were subjected to metagenomic analysis.
For surface sterilization, they were immersed in 70% ethanol for 2 min. following by 5 min. in 5% sodium hypochlorite and then rinsed three times (1 min. each) with sterile distilled water. After drying on sterile filter paper, the fruiting bodies were cut and 100 mg samples of glebal tissue homogenized in liquid nitrogen were taken for DNA isolation using Plant DNA Mini Kit (Syngen Biotech, Poland), following the manufacturer’s protocol. Quality of the DNA was checked on the basis of electrophoregram in 1% TAE-agarose gel. Isolated DNA was stored at – 20 °C. The gleba was also collected for the cultivation of fungi and bacteria for further analysis.
Bacterial and fungal taxa were identified based on 16S rRNA gene sequence and the ITS1 rDNA region, respectively (Haas et al. 2011; Kõljalg et al. 2013; Martin and Rygiewicz, 2005; Thijs et al. 2017). Since no representatives of Archaebacteria were identified in the present study, we use the term “bacteria “ to represent Prokaryote throughout the text. The V3–V4 variable region of 16S rDNA was amplified in the total volume of 25 µl using 5 µl of 1 µM primers 341F and 785R (Kõljalg et al. 2013) and the ITS1 region using primers ITS1FI2 (Schmidt et al. 2013) and 5.8S (Vilgalys and Hester 1990) (Table 1). PCR reactions were performed using 2.5 µl (5 ng/µl) of DNA template and 12.5 µl of 2xKAPA HiFi Hot Start Ready Mix (Kapa Biosystems) and following PCR. The same standard conditions were used for both ITS and 16S: initial denaturation at 95 °C for 3 min, followed by 25 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s with a final extension step at 72 °C for 5 min (White et al. 1990; Thijs et al. 2017). Sequencing of the PCR products was done by Genomed (Warsaw, Poland) using Illumina MiSeq Instrument and pair-end (2 × 250 bp) mode with V2Illumina kit (Balint et al. 2014). Control reaction was performed without DNA added. A preliminary sequence analysis was performed using MiSeq Reporter (MSR) v2.6 program and full comparative analysis using QIIME (Caporaso et al. 2010) with the GreenGenes v13-8 database as a reference for bacteria and UNITE v7 reference database for fungi. Singletons (OTUs represented by a single sequence) were excluded from data analysis (minimal OTU count = 10) and OTUs were picked with sequence identity criteria of 97%.
Cultivation of fungi and bacteria from gleba of T. aestivum fruiting bodies
To cultivate fungi inhabiting gleba of the truffles, gleba samples collected as described above were transferred to Petri dishes containing PDA medium (Potato Dextrose Agar) supplemented with chloramphenicol (0.01%), to inhibit bacterial growth and Bengal rose (0.005%), to inhibit overgrowth by rapidly growing molds and to facilitate isolation of slow-growing fungi (King et al. 1979) and cultivated at 28 °C. Single fungal colonies were sub-cultured on separate Petri dishes containing PDA medium to obtain pure isolates for further studies. Pure culture isolates have been deposited at the culture collection of the Laboratory of Fungal Glycobiology IBB PAS, Warsaw, Poland.
For DNA extraction, mycelia were cultivated in PDB medium (Potato Dextrose Broth) at 28 °C on a rotary shaker (250 rpm) in 250 ml shake flasks containing 100 ml of medium. DNA was isolated using the Wizard Genomic DNA Purification kit (Promega, Mannheim, Germany). Extracted DNA was used as the template for amplification of the ITS region using universal primers ITS1 and ITS4 (White et al. 1990) and a standard PCR protocol (Martin and Rygiewicz 2005). Amplified products were separated by electrophoresis on an agarose gel, isolated from the gel by Qiaex II Gel extraction kit (Qiagen) and sequenced. DNA sequences were analyzed using the NCBI database and the BLAST algorithm (Altschul et al.1997).
Identified species were classified according to the MycoBank fungal database (nomenclature and species bank) https://www.mycobank.org.
For isolation of cultivable bacteria, gleba samples were homogenized in sterile 0.85% NaCl and plated on Tryptone Soy Agar (TSA) (Sigma–Aldrich) as described (Barbieri et al. 2005). Single colonies were sub-cultured on separate Petri dishes containing TSA medium to obtain pure isolates.
DNA was extracted from bacterial colonies (220 pure isolates)) resuspended in TES buffer (20 mM Tris, 50 mM EDTA, 150 mM NaCl, pH 7.9) and lysed with lysozyme (5 mg ml−1) (Sigma –Aldrich) and incubated at 37 °C for 1 h. DNA was extracted lysed with lysozyme using standard phenol extraction as described by Barbieri et al. (2005). Isolated DNA was used as the template for amplifying the DNA between positions 27 and 1492 of bacterial 16S rRNA genes (numbered according to the Escherichia coli rRNA) using primers F27 and R1492 and standard PCR protocol (Frank et al. 2008). DNA sequences were analyzed as above (Altschul et al. 1997). Acc. no. for bacterial strains deposited in NCBI are shown in the Table 5.
Results
Bacterial community inhabiting gleba of T. aestivum
The microbiome of the gleba was determined individually for six truffle fruiting bodies. Illumina sequencing detected 1,012,979 classified bacterial reads for six fruiting bodies (Table 2). 16% of the classified reads (163,937 reads) has been assigned to 20 bacterial OTUs (Table 1S-6S) that belonged to 9 bacterial phyla, 13 classes, 14 orders and 22 families.
Analysis showed that Proteobacteria dominated the bacterial community (Table 2 and Table 1S-6S), with 88.48–96.80% of all identified sequences belonging to the phylum Proteobacteria. The second most common phylum was Bacterioidetes: 2.67–10.13% sequences were assigned to this phylum in three fruiting bodies (1, 4 and 6). Six more phyla were only modestly represented: Acidobacteria, Chloroflexi, Firmicutes, Planctomycetes, Verrucomicrobia, and TM7 a candidate phylum a close relative of the Chloroflexi.
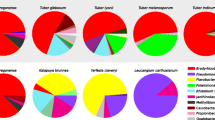
The microbiome of specimen 6 was different from the others, which was well visible at the class level (Fig. 2 and Tables 1S–6S). In this sample, only 45.51% of the sequences were assigned to alpha-Proteobacteria while in the other fruiting bodies this proportion varied from 74.67 to 95.3%. In specimen 6, 31.02% of the sequences belonged to gamma-Proteobacteria, which were represented by a significant fraction of sequences (10.09%) in only one other specimen, no.4 (Fig. 2 and Tables 1S–6S).
Bacterial diversity at class level in gleba of six fruiting bodies of T. aestivum. Illumina sequencing of variable region V3-V4 of 16S rDNA amplified from total gleba DNA was used to identify bacterial classes. (For details see Table 1S-6S). Percentage of qualified reads assigned to each bacterial class is shown
Class alpha-Proteobacteria was represented by five families mostly from the order Rhizobiales. Family Bradyrhizobiaceae from this order was the most common in specimen 2 where 94.31% of sequences were assigned to this family, while in specimen 6 only 41.11% (Tables 1S–6S). In the specimen, 6 family Pseudomonadaceae belonging to gamma-Proteobacteria was more common (30.61%) than in any other specimens, where the percentage of sequences assigned to Pseudomonadaceae varied from 0 to 8.73%.
In total, 20 strains were identified to the genus level. Genus Acidivorax belonging to beta-Proteobacteria, order Burkholderiales, family Comamonadaceae was identified in all six specimens. Family Comamonadaceae was also represented by genera Variovorax, found in four specimens, and Roseateles found only in specimen 4. Eight genera of bacteria were found only in a single specimen (Tables 1S–6S).
Fungal community in T. aestivum gleba
To characterize the fungal microbiome associated with the gleba of T. aestivum fruiting bodies we analyzed the ITS1 (internal transcribed spacer) region of rDNA in total DNA individually for six truffle specimens.
A total of 599,375 classified reads were obtained from the six specimens (Table 3), a vast majority (from 99.74 to 99.99%) was assigned to T. aestivum OTU. However, sequences assigned to other fungal OTUs from three phyla (0.03% Ascomycota other than Tuber, 0.03% Basidiomycota and 0.006% Mucoromycota) were also detected.
In all, six fungal species were found to reside inside the examined fruiting bodies of T. aestivum (Fig. 3 and Table 7S).
The largest variety of fungal sequences was found in specimen 1(five additional species) and in specimens 2 and 4 (three species each). In specimens 5 and 6 two fungal species were identified. No alien fungi were identified in specimen 3.
Sequences representing Trichoderma neokoningi Samuels & Soberanisan and Malassezia restricta E. Guého, J. Guillot & Midgley were found in five (specimens 1, 2, 4, 5, 6) and four (specimens 1, 2, 5, 6) fruiting bodies, respectively, and those representing Mycosphaerella tassiana (De Not.) Johanson, in two specimens (specimen 1 and 2) and Umbelopsis isabellina (Oudem.) W. Gams (specimen 2) and Sphaerodes fimicola (E.C. Hansen) P.F. Cannon & D. Hawksw (specimen 4) – in a single specimen only.
Cultivable fungi and bacteria inhabiting T. aestivum gleba
For a complete picture of the fungal community inhabiting the fruiting bodies of T. aestivum the alien fungi were grown directly from the gleba samples. Ten cultivable fungal species were identified with identity from 83 to 99% (Table 4) and an additional strain could be identified only to the family level. The obtained sequence of Phlebia (Table 4) was in 99% identical to P. rufa and P. radiate, as well.
Most of the identified fungi belonged to the phylum Basidiomycota, four and two representing the Polyporales and Agaricales orders, respectively, and one belonged to order Gloeophyllales. The other four species were assigned to the phyla Ascomycota and Mucoromycota. Notably, none of these isolates corresponded to the fungal species identified above by metagenomics sequencing.
The T. aestivum gleba was also analyzed in a similar manner for cultivable bacteria. We used Tryptone Soy Agar (TSA) medium which has been used earlier to study cultivable bacteria from the ascocarps of T. borchii and T. magnatum (Barbieri et al. 2000, 2005, 2007). Cultivable bacteria were identified with identity at least 96% (Table 5), one obtained sequence was identical in 97% with Bacillus simplex and Bacillus huizhouensis 16S sequence. The same level of identity was obtained for the second sequence identified as B. toyoensis and B. thuringensis 16S sequences.
All the specimens were colonized by different strains of genus Bacillus, family Bacillaceae, order Bacillales, class Bacilli, phylum Firmicutes (Table 5 and Fig. 4). The largest number of bacterial colonies were grown from specimen 6 and the lowest from specimen 2. Bacillaceae family was not assigned to any of the samples in the metagenomics studies.
These results were fundamentally different from those described above for the metagenomics approach. There, order Bacillales was only represented by Staphylococcus aureus, family Staphylococcaceae (in specimens 1, 2 and 5) and no representative of family Bacillaceae was identified.
Discussion
Bacterial microbiome
Truffles host a diverse microbiome inside their fruiting bodies, comprising bacteria (Antony-Babu et al. 2014; Barbieri et al. 2005, 2007, 2016; Benucci and Bonito 2016; Citterio et al. 1995; Gryndler et al. 2013; Pacioni 1990; Sbrana et al. 2002; Splivallo et al. 2015), yeasts (Buzzini et al. 2005) filamentous fungi (Pacioni et al. 2007) and viruses (Stielow and Menzel 2010; Stielow et al. 2011a, b, 2012).
The first study of the bacterial microbiome of T. borchii revealed that most 16S rDNA sequences belonged to alpha-Proteobacteria, with over 97% of them assigned to Rhizobium and Bradyrhizobium spp. (Barbieri et al. 2005).
The factors which regulate microorganisms’ association with the truffle gleba are unknown. Antony-Babu et al. (2014) hypothesized that particular bacterial communities could participate in the truffle development and are selected to the microbiome of the ascocarp during its maturation. A detailed study by that group has shown that the gleba microbiome of T. melanosporum evolves during ascocarp formation. Differences in the microbiome composition have also been shown for the maturing ascocarp of T. magnatum, where alpha-Proteobacteria mainly from the genera Sinorhizobium, Rhizobium and Bradyrhizobium were identified (Barbieri et al. 2007).
The bacterial communities of several Tuber species (T. oregonense Trappe, Bonito & Rawlinson, T. gibbosum Harkn., T. lyonii Butters, T. melanosporum, T. indicum Cooke & Massee) from different geographical regions (Europe, USA and Asia) revealed significant differences in their OTUs but in all specimens sequences from Bradyrhizobium spp. alpha-Proteobacteria were dominant (Benucci and Bonito 2016).
Results of this study, concerning T. aestivum ideally match the earlier data for other Tuber species. We found out that the bacteria of family Bradyrhizobiaceae were the most abundant inhabitants in all examined specimens of T. aestivum. It seems the bacteria belonging to this family also dominate in case of all truffle species. Additionally, the fruiting bodies of T. aestivum (this study), T. borchii (Barbieri et al. 2005) and T. magnatum (Barbieri et al. 2007) all contained such bacteria as Bosea spp. (Bradyrhizobiaceae), but also beta-Proteobacteria, (Variovorax spp.) and gamma-Proteobacteria, (Pseudomonas spp.). Pseudomonas has also been isolated by Citterio et al. (1995) from the fruiting bodies of T. magnatum, T. borchii and T. maculatum Vittad. together with Staphylococcus belonging to the phylum Firmicutes (class Bacilli). We also found Staphylococcus spp. in three specimens of T. aestivum. On the other hand, Staphylococcus spp. seems to be widely represented in the microbiome of fungal fruiting bodies since it was found also in fruiting bodies of forest mushrooms Agaricales, Boletales, Russulales and Cantharellales all from Basidiomycota phylum (Pent et al. 2017).
The overall similarity of the bacterial communities inhabiting different Tuber species suggests that, regardless of the species, the truffle fruiting body creates a specific habitat for a core bacterial microbiome (Splivallo et al. 2015).
The core bacterial microbiome is supplemented specifically depending on the truffle species and the environment. Thus, Acidovorax spp. (beta-Proteobacteria) was identified in all specimens of T. aestivum (this study), but not in T. borchii or T. magnatum ascocarps (Barbieri et al. 2005, 2007). Furthermore, to the best of our knowledge, another beta-Proteobacteria, Cupriavidus sp., found in four of our specimens of T. aestivum has not been detected so far in any of the truffle species studied.
Our cultivation-based approach identified six strains of Bacillus not represented in the molecular analysis. However, there were general similarities between the two approaches, both of which found the highest bacterial diversity in specimen 6 and the lowest in specimen 2.
On the other hand, Barbieri et al. (2005) noticed that many species of bacteria resist cultivation because of their interdependencies with other microbes or because of the lack of knowledge concerning their specific growth requirements. Given that a large portion of environmental bacteria has not been cultured yet, the bacteria isolated thus far could represent only a fraction of the entire natural bacterial community associated with truffles.
Fungal microbiome
The truffle fruiting bodies have been shown to host not only bacteria but also yeasts and filamentous fungi (Buzzini et al. 2005; Pacioni et al. 2007).
Pacioni et al. (2007) analyzed guest cultivable fungi from the gleba of ten Tuber spp. They failed to obtain any mycelial isolates from T. aestivum despite having studied nine specimens but did isolate several filamentous fungi from other Tuber species. In this study, we identified eleven cultivable fungi from the gleba of T. aestivum. Four strains were assigned to the order Polyporales not represented in any other Tuber species described by Pacioni et al. (2007). In contrast, the culture-independent molecular studies of the fungal microbiome of T. aestivum gleba identified Trichoderma neokoningi from the order Hypocreales and this order was represented in other Tuber species although not by Trichoderma (Pacioni et al. 2007).
Identified fungi T. neokoningi, Mycosphaerella tassiana and Umbelopsis isabellina are common soil and plant root inhabiting species while Sphaerodes fimicola was reported to be a coprophilous fungus found in deer fecal samples (Caretta and Piontelli 1996). It is known that truffle spores are passively dispersed by animals (Laessoe and Hansen 2007; Ori et al. 2018). Together with truffles, they could also potentially distribute other fungi as S. fimicola or Malassezia restricta. However, we do not know if the soil fungi play a role in the development of T. aestivum fruiting body or they are just present in the soil because of other reason such as the spread of feces.
The data on the fungal microbiome of Tuber spp. fruiting bodies is limited but more information regarding the fungi inhabiting the soil and root systems of plants infected by truffle are available (Benucci et al. 2011; Leonardi et al. 2013; Li et al. 2017; Mello et al. 2010, 2011; Napoli et al. 2010; Pruett et al. 2008; Zacchi et al. 2003). As could be expected, some such fungi are typical mycorrhizal microorganisms (Benucci et al. 2011; Leonardi et al. 2013; Li et al. 2017) characteristic for each tree and its surrounding (Beckers et al. 2017; Murat et al. 2005). The two localities where truffles were harvested differed in the type of stand. Specimens 1–3 were sampled in the broadleaved forest with Quercus petraea, Acer pseudoplatanus, and Carpinus betulus and specimens 4–6 come from the thicket with Carpinus betulus, Acer campestre and Populus tremula. In addition, the microbial community in the vicinity of Tuber ectomycorrhize could be modulated by the presence of the Tuber mycelia (Leonardi et al. 2013). Those authors observed that the dominance of Tuber mycelia resulted in a lower diversity and abundance of endophytic pathogenic fungi and demonstrated that the bacterial diversity of the ectomycorhizosphere soil was significantly lower than that of more distant soil. This phenomenon could be a plausible explanation of why the community of bacteria and fungi in the Tuber gleba is even more limited. However, since we did not investigate microorganisms inhabiting soil at our location, it is only our supposition.
Another significant phenomenon is that the Tuber fruiting bodies selectively recruit microorganisms to their gleba, but the criteria and mechanisms of this recruitment remain unknown. Notably, we found that the microbiomes of individual fruiting bodies differed from one another markedly, although all the specimens were at the same stage of maturation.
The role of the bacterial and fungal communities residing inside fruiting bodies is controversial but it has been proposed that the bacteria could participate in the development and maturation of truffles (Antony-Babu et al. 2014; Barbieri et al. 2010; Buzzini et al. 2005; Splivallo et al. 2015; Splivallo and Ebeler 2015; Vahdatzadeh et al. 2015—review). It is also assumed that they contribute to the characteristic flavor, which may vary quite substantially between specimens (Vahdatzadeh et al. 2015 – review).
The same role could be attributed to fungi. Sporobolomyces roseus found in our samples of T. aestivum gleba and previously reported as a grape-associated fungus could produce aromatic compounds typical for the red wine aroma (Verginer et al. 2010). Fungi could also play a protective role in producing antibacterial metabolites, which has been shown for Trichopezizella nidulus (J.C. Schmidt & Kunze) Raitv. found in Tuber nitidum Vittad. and Talaromyces wortmannii (Klöcker) C.R. Benj. found in T. rufum Picco (Bara et al. 2003; Pacioni et al. 2007; Thines et al. 1998).
In summary, our study has shown that individual fruiting bodies of T. aestivum collected from two areas, representing the same stage of maturity, possessed different microbiomes. These differences may result from the diverse plant community of forest floor environment (Hilszczańska et al. 2018) and, therefore, influence differences in microbiomes. In general, the core bacterial microbiome of the studied gleba from T. aestivum was similar to the bacterial communities of the other Tuber species (Benucci and Bonito 2016).
Most studies focused on bacterial communities and their role in truffle species. Knowledge about the fungal microbiome and its functions in Tuber spp. is very limited. The fungal microbiome should be examined in detail, and the test should be carried out not only for T. aestivum but also for other Tuber spp.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Antony-Babu S, Deveau A, Van Nostrand JD, Zhou J, Le Tacon F, Robin C, Frey-Klett P, Uroz S (2014) Black truffle - associated bacterial communities during the development and maturation of Tuber melanosporum ascocarps and putative functional roles. Environm Microbiol 16:2831–2847. https://doi.org/10.1111/1462-2920.12294
Balint M, Schidt PA, Sharma R, Thines M, Schmidt I (2014) An Illumina metabarcoding pipeline for fungi. Ecol Evol 4:2642–2653. https://doi.org/10.1002/ece3.1107
Bara R, Aly AH, Pretsch A, Wray V, Wang B, Proksch P, Debbab A (2013) Antibiotically active metabolites from Talaromyces wortmannii, an endophyte of Aloe vera. J Antibiotics 66:491–493
Barbieri E, Bertini L, Rossi I, Ceccaroli P, Saltarelli R, Guidi C, Zambonelli A, Stocchi V (2005) New evidence for bacterial diversity in the ascoma of the ectomycorrhizal fungus Tuber borchii Vittad. FEMS Microbiol Lett 247:23–35. https://doi.org/10.1016/j.femsle.2005.04.027
Barbieri E, Ceccaroli P, Agostini D, Zeppa S D, Gioacchini A M, Stocchi V. (2016) Truffle-associated bacteria: Extrapolation from diversity to function. In: Zambonelli A, Iotti M, Murat C (eds) True truffle (Tuber spp.) in the world. Soil ecology, systematics and biochemistry. Springer, pp 301–318
Barbieri E, Ceccaroli P, Saltarelli R, Guidi Ch, Potenza L, Basaglia M, Fontana F, Baldan E, Casella S, Ryahi O, Zambonelli A, Stocchi V (2010) New evidence for nitrogen fixation within the Italian white truffle Tuber magnatum. Fungal Biol 114:936–942. https://doi.org/10.1016/j.funbio.2010.09.001
Barbieri E, Guidi C, Bertaux J, Frey-Klett P, Garbaye J, Ceccaroli P, Saltarelli R, Zambonelli A, Stocchi V (2007) Occurrence and diversity of bacterial communities in Tuber magnatum during truffle maturation. Environm Microbiol 9:2234–2246. https://doi.org/10.1111/j.1462-2920.2007.01338.x
Barbieri E, Potenza L, Rossi I, Sisti D, Giomaro G, Rossetti S, Beimfohr C, Stocchi V (2000) Phylogenetic characterization and in situ detection of a Cytophaga-Flexibacter-Bacteroides phylogroup bacterium in Tuber borchii Vittad. ectomycorrhizal mycelium. Appl Environm Microbiol 66:5035–5042. https://doi.org/10.1128/aem.66.11.5035-5042.2000
Beckers B, Op De Beeck M, Weyens N, Boerjan W, Vangronsveld J (2017) Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome. https://doi.org/10.1186/s40168-017-0241-2
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2013) GenBank. Nucleic Acids Res 41(D1):D36–D42
Benucci GMN, Bonito GM (2016) The truffle microbiome: Species and geography effects on bacteria associated with fruiting bodies of hypogenous Pezizales. Microb Ecol 72:4–8
Benucci GMN, Raggi L, Albertini E, Grebenc T, Bencivenga M, Falcinelli M, Di Massimo G (2011) Ectomycorrhizal communities in a productive Tuber aestivum Vittad. orchard: composition, host influence and species replacement. FEMS Microbiol Ecol 76:170–184. https://doi.org/10.1111/j.1574-6941.2010.01039.x
Büntgen U, Tegel W, Egli S, Stobbe U, Sproll L, Stenseth NC (2011) Truffles and climate change. Front Ecol Environ 9:150e151
Büntgen U, Bagi I, Fekete O, Molinier V, Peter M, Splivallo R, Vahdatzadeh M, Richard F, Murat C, Tegel W, Stobbe U, Martínez-Peña F, Sproll L, Hülsmann L, Nievergelt D, Meier B, Egli S (2017) New insights into the complex relationship between weight and maturity of Burgundy truffles (Tuber aestivum). PLoS ONE 12:e0170375. https://doi.org/10.1371/journal.pone.0170375
Buzzini P, Gasparetti C, Turchetti B, Cramarossa MR, Vaughan-Martini A, Martini A, Pagnoni UM, Forti L (2005) Production of volatile organic compounds (VOCs) by yeasts isolated from the ascocarps of black (Tuber melanosporum Vitt.) and white (Tuber magnatum Pico) truffles. Arch Microbiol 184:187–193. https://doi.org/10.1007/s00203-005-0043-y
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez-Pena A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig J, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Caretta G, Piontelli E (1996) Coprophilous fungi from confined deers in Pavia (Lombardia, Italy). Boletin Micologico 11:41–50
Citterio B, Cardoni P, Potenza L, Amicucci A, Stocchi V, Gola G, Nuti M (1995) Isolation of bacteria from sporocarps of Tuber magnatum Pico, Tuber borchii Vitt. and Tuber maculatum Vitt. In: Stocchi V, Bonfante P, Nuti M (eds) Biotechnology of ectomycorrhizae. Plenum Press, New York, pp 241–248
Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ (2008) Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environm Microbiol 74:2461–2470. https://doi.org/10.1128/AEM.02272-07
Gryndler M, Soukupova L, Hrselova H, Gryndlerova H, Borovicka J, Streiblova E, Jansa J (2013) A quest for indigenous truffle helper prokaryotes: Tuber aestivum-associative prokaryotes. Environm Microbiol Rep 5:346–352. https://doi.org/10.1111/1758-2229.12014
Hall IR, Yun W, Amicucci A (2003) Cultivation of edible ectomycorrhizal mushrooms. Trends Biotechnol 21:433–438. https://doi.org/10.1016/S0167-7799(03)00204-X
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodargren E, Methe B, DeSantis TZ, Petrosino JF, Knight R, Birren BW (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. https://doi.org/10.1101/gr.112730.110
Hilszczańska D, Sierota Z, Palenzona M (2008) New Tuber species found in Poland. Mycorrhiza 18:223–226. https://doi.org/10.1007/s00572-008-0175-4
Hilszczańska D, Rosa-Gruszecka A, Szmidla H (2014) Characteristic of Tuber spp. localities in natural stands with emphasis on plant species composition. Acta Mycol 49:267–277. https://doi.org/10.5586/am.2014.024
Hilszczańska D, Siebyła M, Horak J, Król M, Podsadni P, Steckiewicz P, Bamburowicz-Klimkowska M, Szutowski M, Turło J (2016) Comparison of chemical composition in Tuber aestivum Vittad. of different geographical origin. Chem Biodiversity 13:1617–1629. https://doi.org/10.1002/cbdv.201600041
Hilszczańska D, Rosa-Gruszecka A, Gawryś R (2018) Horak J (2018) Effect of soil properties and vegetation characteristics in determining the frequency of Burgundy truffle fruiting bodies in Southern Poland. Écoscience 10(1080/11956860):1530327
King AD Jr, Hocking AD, Pitt JI (1979) Dichloran-rose Bengal medium for enumeration and isolation of molds from foods. Appl Environm Microbiol 37:959–964
Koljalg U, Nilson RH, Abarenkov K, Tedersoo L, Taylor AF, Bahram M, Bates ST, BrunsTD Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martí MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson KH (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277. https://doi.org/10.1111/mec.12481
Laessoe T, Hansen K (2007) Truffle trouble: what happened to the Tuberales? Mycol Res 111:1075–1099
Leonardi M, Iotti M, Oddis M, Lalli G, Pacioni G, Leonardi P, Maccherini S, Perini C, Salerni E, Zambonelli A (2013) Assessment of ectomycorrhizal fungal communities in the natural habitats of Tuber magnatum (Ascomycota, Pezizales). Mycorrhiza 23:349–358. https://doi.org/10.1007/s00572-012-0474-7
Li Q, Zhao J, Xiong C, Li X, Chen Z, Li P, Huang W (2017) Tuber indicum shapes the microbial communities of ectomycorhizosphere soil and ectomycorrhizae of an indigenous tree (Pinus armandii). PLoS ONE 12(4):e0175720. https://doi.org/10.1371/journal.pone.0175720
Martin KJ, Rygiewicz PT (2005) Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol 5:28. https://doi.org/10.1186/1471-2180-5-28
Mello A, Miozzi L, Vizzini A, Napoli C, Kowalchuk G, Bonfante P (2010) Bacterial and fungal communities associated with Tuber magnatum-productive niches. Plant Biosystems 144:323–332. https://doi.org/10.1080/11263500903374724
Mello A, Murat C, Bonfante P (2006) Truffles: much more than a prized and local fungal delicacy. FEMS Microbiol Lett 260:1–8. https://doi.org/10.1111/j.1574-6968.2006.00252.x
Mello A, Napoli C, Murat C, Marceddu G, Bonfante P (2011) ITS-1 versus ITS-2 pyrosequencing: a comparison of fungal population in truffle grounds. Mycologia 130:1184–1193. https://doi.org/10.3852/11-027
Mello A, Ding GC, Piceno YM, Napoli C, Tom LM, DeSantis TZ, Andersen GL, Smalla K, Bonfante P (2013) Truffle brules have an impact on the diversity of soil bacterial communities. PLoS ONE 8(4):e61945. https://doi.org/10.1371/journal.pone.0061945
Mello A, Zampieri E, Zambonelli A. (2017) Truffle Ecology. In: Varma A, Prased R, Tuteja N. (eds) Genetic diversity, soil interactions and functioning in mycorrhiza. Function, diversity, state of art. Springer Nature, Switzerland, pp 231–253. 10.1007/978–3–319–53064–2
Murat C, Vizzini A, Bonfante P, Mello A (2005) Morphological and molecular typing of the below-ground fungal community in a natural Tuber magnatum truffle-ground. FEMS Microbiol Lett 245:307–313. https://doi.org/10.1016/j.femsle.2005.03.019
Napoli C, Mello A, Borra A, Vizzini A, Sourzat P, Bonfante P (2010) Tuber melanosporum, when dominant, affects fungal dynamics in truffle grounds. New Phytol 185:237–247. https://doi.org/10.1111/j.1469-8137.2009.03053.x
Nazzaro F, Fratianni F, Picariello G, Coppola R, Reale A, Luccia AD (2007) Evaluation of gamma rays influence on some biochemical and microbiological aspects in black truffles. Food Chem 103:344–354. https://doi.org/10.1016/j.foodchem.2006.07.067
Ori F, Trappe J, Leonardi M, Iotti M, Pacioni G (2018) Crested porcupines (Hystrix cristata): mycophagist spore dispersers of the ectomycorrhizal truffle Tuber aestivum. Micorrhiza 28:561–565. https://doi.org/10.1007/s00572-018-0840-1
Pacioni G (1990) Scanning electron microscopy of Tuber sporocarps and associated bacteria. Mycol Res 94:1086–1089. https://doi.org/10.1016/S0953-7562(09)81338-5
Pacioni G, Leonardi M. (2016) Truffle-inhabiting fungi. In: Zambonelli A, Iotti M, Murat C. (eds) True truffle (Tuber spp.) in the world. Soil ecology, systematics and biochemistry. Springer, pp 283–299.
Pacioni G, Leonardi M, Aimola P, Ragnelli AM, Rubini A, Paolocci F (2007) Isolation and characterization of some mycelia inhabiting Tuber ascomata. Mycol Res 111:1450–1460. https://doi.org/10.1016/j.mycres.2007.08.016
Pacioni G, Ragnelli AM, Miranda M (1995) Truffles development and interactions with the biotic environment: molecular aspects. In: Stocchi V, Bonfante P, Nuti M (eds) Biotechnology of ectomycorrhizae. Plenum Press, New York, pp 213–227
Pent M, Poldmaa K, Bahram M (2017) Bacterial communities in boreal forest mushrooms are shaped both by soil parameters and host identity. Frontiers in Microbiol. https://doi.org/10.3389/fmicb.2017.00836
Pieroni A (2016) The changing ethnoecological cobweb of white truffle (Tuber magnatum Pico) gatherers in South Piedmont. J Ethnobiol Ethnomed, NW Italy. https://doi.org/10.1186/s13002-016-0088-9
Pruett G, Bruhn J, Mihail J (2008) Temporal dynamics of ectomycorrhizal community composition on root systems of oak seedlings infected with Burgundy truffle. Mycol Res 112:1344–1354. https://doi.org/10.1016/j.mycres.2008.06.005
Rivera CS, Blanco D, Oria R, Venturini ME (2010) Diversity of culturable microorganisms and occurrence of Listeria monocytogenes and Salmonella spp. in Tuber aestivum and Tuber melanosporum ascocarps. Food Microbiol 27:286–293. https://doi.org/10.1016/j.fm.2009.11.001
Salerni E, Iotti M, Leonardi P, Gardin L, D'Aguanno M, Perini C, Pacioni P, Zambonelli A (2014) Effects of soil tillage on Tuber magnatum development in natural truffières. Mycorrhiza. 1:79–87. https://doi.org/10.1007/s00572-013-0543-6
Sbrana C, Agnolucci M, Bedini S, Lepera A, Toffanin A, Giovannetti M, Nuti MP (2002) Diversity of culturable bacterial populations associated to Tuber borchii ectomycorrhizas and their activity on T. borchii mycelial growth. FEMS Microbiol Lett 211:195–201. https://doi.org/10.1111/j.1574-6968.2002.tb11224.x
Schmidt PA, Balint M, Greshake B, Bandow C, Rombke J, Schmitt I (2013) Illumina metabarcoding of a soil fungal community. Soil Biol Biochem 65:128–132. https://doi.org/10.1016/j.soilbio.2013.05.014
Splivallo R, Deveau A, Valdez N, Kirchhoff N, Frey-Klett P, Karlovsky P (2015) Bacteria associated with truffle-fruiting bodies contribute to truffle aroma. Environm Microbiol 17:2647–2660. https://doi.org/10.1111/1462-2920.12521
Splivallo R, Ebeler SE (2015) Sulfur volatiles of microbial origin are key contributors to human-sensed truffle aroma. Appl Microbiol Biotechnol 99:2583–2592. https://doi.org/10.1007/s00253-014-6360-9
Stielow B, Menzel W (2010) Complete nucleotide sequence of TaV1, a novel totivirus isolated from a black truffle ascocarp (Tuber aestivum Vittad.). Arch Virol 155:2075–2078. https://doi.org/10.1007/s00705-010-0824-8
Stielow B, Klenk HP, Menzel W (2011) Complete genome sequence of the first endornavirus from the ascocarp of the ectomycorrhizal fungus Tuber aestivum Vittad. Arch Virol 156:343–345. https://doi.org/10.1007/s00705-010-0875-x
Stielow JB, Klenk HP, Winter S, Menzel W (2011) A novel Tuber aestivum (Vittad) mitovirus. Arch Virol 156:1107–1110. https://doi.org/10.1007/s00705-011-0998-8
Stielow JB, Bratek Z, Klenk HP, Winter S, Menzel W (2012) A novel mitovirus from the hypogeous ectomycorrhizal fungus Tuber excavatum. Arch Virol 157:787–790. https://doi.org/10.1007/s00705-012-1228-8
Stobbe U, Egli S, Tegel W, Pete M, Sproll L, Buntgen U (2013) Potential and limitations of Burgundy truffle cultivation. Appl Microbiol Biotechnol 97:5215–5224. https://doi.org/10.1007/s00253-013-4956-0
Thijs S, De Beck M, Backers B, Truyens S, Stevens V, Van Hamme JD, Weyens N, Vangronsveld J (2017) Comparative evaluation of four bacteria-specific primer pairs for 16S rRNA gene surveys. Frontiers in Microbiol. https://doi.org/10.3389/fmicb.2017.00494
Thines E, Anke H, Sterner O (1998) Trichoflectin, a bioactive azaphilone from the ascomycete Trichopezizella nidulus. J Nat Prod 61:306–308
Vahdatzadeh M, Deveau A, Splivallo R (2015) The role of the microbiome of truffles in aroma formation: a meta-analysis approach. Appl Environm Microbiol l 81:6946–6952. https://doi.org/10.1128/AEM.01098-15
Verginer M, Leitner E, Berg G (2010) Production of volatile metabolites by grape-associated microorganisms. J Agric Food Chem 58:8344–8350. https://doi.org/10.1021/jf100393w
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 17:4238–4246
Wang X, Benucci GMN, Xie X, Bonito G, Leisola M, Liu P, Shamekh S (2013) Morphological, mycorrhizal and molecular characterization of Finnish truffles belonging to the Tuber anniae species-complex. Fungal Ecol 6:269–280. https://doi.org/10.1016/j.funeco.2013.03.002
Weden Ch, Danell E, Tibell L (2005) Species recognition in the truffle genus Tuber – the synonyms Tuber aestivum and Tuber uncinatum. Environm Microbiol 7:1535–1546. https://doi.org/10.1111/j.1462-2920.2005.00837.x
White TJ, Bruns T, Lee S, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ. White TJ (eds) PCR Protocols: A guide to methods and applications. Academic Press Inc., New York, pp 315–322
Zacchi L, Vaughan-Martini A, Angelini P (2003) Yeast distribution in a truffle field ecosystem. Annals Microbiol 53:275–282
Zambonelli A, Iotti M, Piattioni F (2012) Chinese Tuber aestivum sensu lato in Europe. Open Mycol J 6:22–26. https://doi.org/10.2174/1874437001206010022
Zambonelli A, Iotti M, Hall I (2015) Current status of truffle cultivation: recent results and future perspectives Micol. Ital 44:31–40. https://doi.org/10.6092/issn.2465-311X/559331
Zarivi O, Cesare P, Ragnelli AM, Aimola P, Leonardi M, Bonfigli A, Colafarina S, Poma AM, Miranda M, Pacioni G (2015) Validation of reference genes for quantitative real-time PCR in Perigord black truffle (Tuber melanosporum) developmental stages. Phytochem 116:78–86. https://doi.org/10.1016/j.phytochem.2015.02.024
Acknowledgements
All reviewers are acknowledged for their valuable comments and suggestions, which greatly improved the paper.
Funding
The research was performed within the project financed by the State Forests National Forest Holding [Grant no. OR-271.3.5.2017].
Author information
Authors and Affiliations
Contributions
Conceptualization: J.S.K., J.T., D.H., Methodology: J.S.K., J.T., D.H., Investigation: U.P-L, S.P., E.G., Writing- original draft preparation: J.S.K., Writing-review and editing: J.S.K., Supervision: J.S.K., D.H., J.T., Project administration: J.S.K.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Perlińska-Lenart, U., Piłsyk, S., Gryz, E. et al. Identification of bacteria and fungi inhabiting fruiting bodies of Burgundy truffle (Tuber aestivum Vittad.). Arch Microbiol 202, 2727–2738 (2020). https://doi.org/10.1007/s00203-020-02002-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-020-02002-x