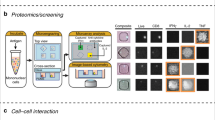
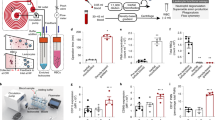
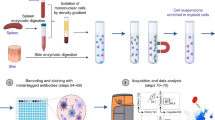
Abstract
Characterization of immune responses is currently hampered by the lack of systems enabling quantitative and dynamic phenotypic characterization of individual cells and, in particular, analysis of secreted proteins such as cytokines and antibodies. We recently developed a simple and robust microfluidic platform, DropMap, to measure simultaneously the kinetics of secretion and other cellular characteristics, including endocytosis activity, viability and expression of cell-surface markers, from tens of thousands of single immune cells. Single cells are compartmentalized in 50-pL droplets and analyzed using fluorescence microscopy combined with an immunoassay based on fluorescence relocation to paramagnetic nanoparticles aligned to form beadlines in a magnetic field. The protocol typically takes 8–10 h after preparation of microfluidic chips and chambers, which can be done in advance. By contrast, enzyme-linked immunospot (ELISPOT), flow cytometry, time-of-flight mass cytometry (CyTOF), and single-cell sequencing enable only end-point measurements and do not enable direct, quantitative measurement of secreted proteins. We illustrate how this system can be used to profile downregulation of tumor necrosis factor-α (TNF-α) secretion by single monocytes in septic shock patients, to study immune responses by measuring rates of cytokine secretion from single T cells, and to measure affinity of antibodies secreted by single B cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are not publicly available due to confidentiality and contractual obligations toward industrial partners of the project in which the data were generated but are available from the corresponding author upon reasonable request.
Code availability
The DropMap MATLAB script is available from the GitHub repository, https://github.com/LCMD-ESPCI/dropmap-analyzer. MATLAB scripts illustrating the main functions performed by the DropCell.exe MATLAB application are available from GitHub repository https://github.com/bioaster/dropcell.git. The installation file for the DropCell.exe MATLAB application, as well as an image dataset that supports/illustrates the findings of this study, are available in the figshare repository, with the identifiers https://figshare.com/articles/DropCell_exe_installer/11336663/1 and https://figshare.com/articles/dropcell_image_data_set/11342426/1, respectively.
Change history
13 January 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41596-021-00492-7
References
Lawson, D. A., Kessenbrock, K., Davis, R. T., Pervolarakis, N. & Werb, Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 20, 1349–1360 (2018).
Potter, S. S. Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 14, 479–492 (2018).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Svensson, V., Vento-Tormo, R. & Teichmann, S. A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599–604 (2018).
Zhang, X. et al. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-Seq systems. Mol. Cell 73, 130–142.e5 (2018).
Battle, A. et al. Genomic variation. Impact of regulatory variation from RNA to protein. Science 347, 664–667 (2015).
Peterson, V. M. et al. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 35, 936–939 (2017).
Shapiro, H. M. Practical Flow Cytometry (Wiley-Liss, 2003).
Bendall, S. C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Becattini, S. et al. Functional heterogeneity of human memory CD4(+) T cell clones primed by pathogens or vaccines. Science 347, 400–406 (2015).
Betts, M. R. et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods 281, 65–78 (2003).
Han, G., Spitzer, M. H., Bendall, S. C., Fantl, W. J. & Nolan, G. P. Metal-isotope-tagged monoclonal antibodies for high-dimensional mass cytometry. Nat. Protoc. 13, 2121–2148 (2018).
Korin, B., Dubovik, T. & Rolls, A. Mass cytometry analysis of immune cells in the brain. Nat. Protoc. 13, 377–391 (2018).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Shahi, P., Kim, S. C., Haliburton, J. R., Gartner, Z. J. & Abate, A. R. Abseq: ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding. Sci. Rep. 7, 44447 (2017).
Czerkinsky, C. C., Nilsson, L. A., Nygren, H., Ouchterlony, O. & Tarkowski, A. A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. J. Immunol. Methods 65, 109–121 (1983).
Kouwenhoven, M. et al. Enzyme-linked immunospot assays provide a sensitive tool for detection of cytokine secretion by monocytes. Clin. Diagn. Lab. Immunol. 8, 1248–1257 (2001).
Schultes, B. C. & Whiteside, T. L. Monitoring of immune responses to CA125 with an IFN-gamma ELISPOT assay. J. Immunol. Methods 279, 1–15 (2003).
Mogensen, T. H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22, 240–273 (2009).
Garcia-Cordero, J. L., Nembrini, C., Stano, A., Hubbell, J. A. & Maerkl, S. J. A high-throughput nanoimmunoassay chip applied to large-scale vaccine adjuvant screening. Integr. Biol. (Camb.) 5, 650–658 (2013).
Han, Q. et al. Polyfunctional responses by human T cells result from sequential release of cytokines. Proc. Natl Acad. Sci. USA 109, 1607–1612 (2012).
Han, Q., Bradshaw, E. M., Nilsson, B., Hafler, D. A. & Love, J. C. Multidimensional analysis of the frequencies and rates of cytokine secretion from single cells by quantitative microengraving. Lab Chip 10, 1391–1400 (2010).
Shirasaki, Y. et al. Real-time single-cell imaging of protein secretion. Sci. Rep. 4, 4736 (2014).
Son, K. J. et al. Microfluidic compartments with sensing microbeads for dynamic monitoring of cytokine and exosome release from single cells. Analyst 141, 679–688 (2016).
Varadarajan, N. et al. A high-throughput single-cell analysis of human CD8(+) T cell functions reveals discordance for cytokine secretion and cytolysis. J. Clin. Invest. 121, 4322–4331 (2011).
Xue, Q. et al. Single-cell multiplexed cytokine profiling of CD19 CAR-T cells reveals a diverse landscape of polyfunctional antigen-specific response. J. ImmunoTher. Cancer 5, 85 (2017).
Xue, Q. et al. Analysis of single-cell cytokine secretion reveals a role for paracrine signaling in coordinating macrophage responses to TLR4 stimulation. Sci. Signal. 8, ra59 (2015).
Yamanaka, Y. J. et al. Cellular barcodes for efficiently profiling single-cell secretory responses by microengraving. Anal. Chem. 84, 10531–10536 (2012).
Seah, Y. F. S., Hu, H. & Merten, C. A. Microfluidic single-cell technology in immunology and antibody screening. Mol. Asp. Med. 59, 47–61 (2018).
Love, J., Ronan, J., Grotenbreg, G., Van Der Veen, A. & Ploegh, H. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat. Biotechnol. 24, 703–707 (2006).
Jin, A. et al. Rapid isolation of antigen-specific antibody-secreting cells using a chip-based immunospot array. Nat. Protoc. 6, 668–676 (2011).
Köster, S. et al. Drop-based microfluidic devices for encapsulation of single cells. Lab Chip 8, 1110–1115 (2008).
Clausell-Tormos, J. et al. Droplet-based microfluidic platforms for the encapsulation and screening of mammalian cells and multicellular organisms. Chem. Biol. 15, 427–437 (2008).
El Debs, B., Utharala, R., Balyasnikova, I. V., Griffiths, A. D. & Merten, C. A. Functional single-cell hybridoma screening using droplet-based microfluidics. Proc. Natl Acad. Sci. 109, 11570–11575 (2012).
Mazutis, L. et al. Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc. 8, 870–891 (2013).
Shembekar, N., Hu, H., Eustace, D. & Merten, C. A. Single-cell droplet microfluidic screening for antibodies specifically binding to target cells. Cell Rep. 22, 2206–2215 (2018).
Chokkalingam, V. et al. Probing cellular heterogeneity in cytokine-secreting immune cells using droplet-based microfluidics. Lab Chip 13, 4740–4744 (2013).
Eyer, K. et al. Single-cell deep phenotyping of IgG-secreting cells for high-resolution immune monitoring. Nat. Biotechnol. 35, 977–982 (2017).
Jorgolli, M. et al. Nanoscale integration of single cell biologics discovery processes using optofluidic manipulation and monitoring. Biotechnol. Bioeng. 116, 2393–2411 (2019).
Mocciaro, A. et al. Light-activated cell identification and sorting (LACIS) for selection of edited clones on a nanofluidic device. Commun. Biol. 1, 41 (2018).
Winters, A. et al. Rapid single B cell antibody discovery using nanopens and structured light. mAbs 11, 1025–1035 (2019).
Konry, T., Dominguez-Villar, M., Baecher-Allan, C., Hafler, D. A. & Yarmush, M. L. Droplet-based microfluidic platforms for single T cell secretion analysis of IL-10 cytokine. Biosens. Bioelectron. 26, 2707–2710 (2011).
Qiu, L. et al. A membrane-anchored aptamer sensor for probing IFNγ secretion by single cells. Chem. Commun. (Camb.) 53, 8066–8069 (2017).
Segaliny, A. I. et al. Functional TCR T cell screening using single-cell droplet microfluidics. Lab Chip 18, 3733–3749 (2018).
Gérard, A. et al. High-throughput single-cell activity-based screening and sequencing of antibodies using droplet microfluidics. Nat. Biotechnol. 38, 715–721 (2020).
Armbruster, D. A. & Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 29(Suppl 1), S49–S52 (2008).
Duffy, D. C., McDonald, J. C., Schueller, O. J. & Whitesides, G. M. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal. Chem. 70, 4974–4984 (1998).
Anna, S. L., Bontoux, N. & Stone, H. A. Formation of dispersions using “flow focusing” in microchannels. Appl. Phys. Lett. 82, 364–366 (2003).
Akhtar, M., van den Driesche, S., Bödecker, A. & Vellekoop, M. J. Long-term storage of droplets on a chip by Parylene AF4 coating of channels. Sens. Actuators B Chem. 255, 3576–3584 (2018).
Di Carlo, D., Aghdam, N. & Lee, L. P. Single-cell enzyme concentrations, kinetics, and inhibition analysis using high-density hydrodynamic cell isolation arrays. Anal. Chem. 78, 4925–4930 (2006).
Jin, S. H., Jeong, H. H., Lee, B., Lee, S. S. & Lee, C. S. A programmable microfluidic static droplet array for droplet generation, transportation, fusion, storage, and retrieval. Lab Chip 15, 3677–3686 (2015).
Liu, C., Liu, J., Gao, D., Ding, M. & Lin, J. M. Fabrication of microwell arrays based on two-dimensional ordered polystyrene microspheres for high-throughput single-cell analysis. Anal. Chem. 82, 9418–9424 (2010).
Ochsner, M. et al. Micro-well arrays for 3D shape control and high resolution analysis of single cells. Lab Chip 7, 1074–1077 (2007).
Schmitz, C. H., Rowat, A. C., Koster, S. & Weitz, D. A. Dropspots: a picoliter array in a microfluidic device. Lab Chip 9, 44–49 (2009).
Duval, F., van Beek, T. A. & Zuilhof, H. Key steps towards the oriented immobilization of antibodies using boronic acids. Analyst 140, 6467–6472 (2015).
Kumar, S., Aaron, J. & Sokolov, K. Directional conjugation of antibodies to nanoparticles for synthesis of multiplexed optical contrast agents with both delivery and targeting moieties. Nat. Protoc. 3, 314–320 (2008).
Saha, B., Evers, T. H. & Prins, M. W. How antibody surface coverage on nanoparticles determines the activity and kinetics of antigen capturing for biosensing. Anal. Chem. 86, 8158–8166 (2014).
Saha, B., Songe, P., Evers, T. H. & Prins, M. W. J. The influence of covalent immobilization conditions on antibody accessibility on nanoparticles. Analyst 142, 4247–4256 (2017).
Sivaram, A. J., Wardiana, A., Howard, C. B., Mahler, S. M. & Thurecht, K. J. Recent Advances in the generation of antibody-nanomaterial conjugates. Adv. Healthc. Mater. 7, 1700607 (2018).
Welch, N. G., Scoble, J. A., Muir, B. W. & Pigram, P. J. Orientation and characterization of immobilized antibodies for improved immunoassays (review). Biointerphases 12, 02d301 (2017).
Dhadge, V. L., Hussain, A., Azevedo, A. M., Aires-Barros, R. & Roque, A. C. Boronic acid-modified magnetic materials for antibody purification. J. R. Soc. Interface 11, 20130875 (2014).
Lin, P. C. et al. Fabrication of oriented antibody-conjugated magnetic nanoprobes and their immunoaffinity application. Anal. Chem. 81, 8774–8782 (2009).
Wang, X., Xia, N. & Liu, L. Boronic acid-based approach for separation and immobilization of glycoproteins and its application in sensing. Int. J. Mol. Sci. 14, 20890–20912 (2013).
Wagner, O. et al. Biocompatible fluorinated polyglycerols for droplet microfluidics as an alternative to PEG-based copolymer surfactants. Lab Chip 16, 65–69 (2016).
Williamson, J. D. & Cox, P. Use of a new buffer in the culture of animal cells. J. Gen. Virol. 2, 309–312 (1968).
Lowe, K. C. Perfluorochemical respiratory gas carriers: benefits to cell culture systems. J. Fluor. Chem. 118, 19–26 (2002).
Holtze, C. et al. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab Chip 8, 1632–1639 (2008).
Mazutis, L. & Griffiths, A. D. Selective droplet coalescence using microfluidic systems. Lab Chip 12, 1800–1806 (2012).
Scott, R. L. The solubility of fluorocarbons. J. Am. Chem. Soc. 70, 4090–4093 (1948).
Simons, J. H. & Linevsky, M. J. The solubility of organic solids in fluorocarbon derivatives. J. Am. Chem. Soc. 74, 4750–4751 (1952).
Qin, D., Xia, Y. & Whitesides, G. M. Soft lithography for micro- and nanoscale patterning. Nat. Protoc. 5, 491–502 (2010).
Eyer, K. et al. The quantitative assessment of the secreted IgG repertoire after recall to evaluate the quality of immunizations. J. Immunol. https://doi.org/10.4049/jimmunol.2000112 (2020).
Raphael, I., Nalawade, S., Eagar, T. N. & Forsthuber, T. G. T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine 74, 5–17 (2015).
Tanaka, A. & Sakaguchi, S. Regulatory T cells in cancer immunotherapy. Cell. Res. 27, 109–118 (2017).
Kang, S., Brown, H. M. & Hwang, S. Direct antiviral mechanisms of interferon-gamma. Immune Netw. 18, e33 (2018).
Mosmann, T. R., Cherwinski, H., Bond, M. W., Giedlin, M. A. & Coffman, R. L. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 136, 2348–2357 (1986).
Wheelock, E. F. Interferon-like virus-inhibitor induced in human leukocytes by phytohemagglutinin. Science 149, 310–311 (1965).
DuPage, M. & Bluestone, J. A. Harnessing the plasticity of CD4(+) T cells to treat immune-mediated disease. Nat. Rev. Immunol. 16, 149–163 (2016).
Cecconi, M., Evans, L., Levy, M. & Rhodes, A. Sepsis and septic shock. Lancet 392, 75–87 (2018).
Gyawali, B., Ramakrishna, K. & Dhamoon, A. S. Sepsis: the evolution in definition, pathophysiology, and management. SAGE Open Med. 7, 2050312119835043 (2019).
Monneret, G. et al. Novel approach in monocyte intracellular TNF measurement: application to sepsis-induced immune alterations. Shock 47, 318–322 (2017).
Shalova, I. N. et al. Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1alpha. Immunity 42, 484–498 (2015).
Kumar, P., Pai, K., Pandey, H. P. & Sundar, S. Study on pinocytosis by monocytes from visceral leishmaniasis patients. Curr. Sci. 83, 631–633 (2002).
Luciani, N., Gazeau, F. & Wilhelm, C. Reactivity of the monocyte/macrophage system to superparamagnetic anionic nanoparticles. J. Mater. Chem. 19, 6373–6380 (2009).
Robert, D. et al. Cell sorting by endocytotic capacity in a microfluidic magnetophoresis device. Lab Chip 11, 1902–1910 (2011).
Acknowledgements
We acknowledge the support of the REALISM study group: HCL: A. Boibieux, J. Davidson, L. Fayolle-Pivot, J. Gatel, C. Genin, A. Gregoire, A. Lepape, A.-C. Lukaszewicz, G. Marcotte, Marie Matray, D. Maucort-Boulch, N. Panel, T. Rimmele, H. Vallin; bioMérieux: S. Blein, K. Brengel-Pesce, E. Cerrato, V. Cheynet, E. Gallet-Gorius, A. Guichard, C. Jourdan, N. Koenig, F. Mallet, B. Meunier, M. Mommert, G. Oriol, C. Schrevel, O. Tabone, J. Yugueros Marcos; Bioaster: J. Becker, F. Bequet, F. Brajon, B. Canard, M. Collus, N. Garcon, I. Gorse, F. Lavocat, K. Louis, J. Moriniere, Y. Mouscaz, L. Noailles, M. Perret, F. Reynier, C. Riffaud, M.-L. Rol, N. Sapay; Sanofi: C. Carre, A. de Monfort, K. Florin, L. Fraisse, I. Fugier, M. L’Azou, S. Payrard, A. Peleraux, L. Quemeneur; ESPCI: S. Toetsch; GSK: T. Ashton, P.J. Gough, S.B. Berger, D. Gardiner, A. MacNamara, A. Raychaudhuri, R. Smylie, L. Tan, C. Tipple. This research project received funding from the French Government through the “Investissement d’Avenir” program (grant no. ANR-10-AIRT-03) and from bioMérieux. K.E. acknowledges generous funding from the ‘The Branco Weiss Fellowship – Society in Science’ and received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 80336). This work also received support from “Institut Pierre-Gilles de Gennes” (laboratoire d’excellence, “Investissements d’Avenir” programs ANR-10-IDEX-0001-02 PSL, ANR-10- EQPX-34 and ANR-10-LABX-31). This work was also supported by BPIFrance under the framework “Programme d’Investissements d’Avenir” (CELLIGO Project). The authors thank the healthy donors and the septic patients who volunteered to donate peripheral blood for these experiments. We also thank M.-N. Unheheuer, H. Laude and B.L. Perlaza for access to the BioResources platform (ICAReB). We thank F. Pène, who collected clinical samples from septic shock patients at the medical intensive care unit of Cochin Hospital (CPP17-053a / 2017-A01134-49). We thank F. Porcheray for critical reading of the manuscript. We further acknowledge the help of P. Canales Herrerias and P. Bruhns for their helpful discussion and supervision of the immunization of mice.
Author information
Authors and Affiliations
Contributions
Y.B., K.E., M.R. and N.A. performed and optimized the experiments described in this protocol, S.D. and G.C. provided the respective MATLAB scripts for data analysis; C.C and T.T. provided the statistical tools. M.M. optimized the boronic acid nanobead functionalization protocol. C.V. and J. Baudry managed the optical bench setup. J.-F.L. supplied the septic clinical samples. K.E., J. Bibette, J. Baudry and A.D.G. developed the DropMap technology38 and contributed to the early-stage definition of the new technology. Y.B. and C.V. extended the DropMap technology to measure low cytokine secretion profiles and to overcome limitations of cell endocytic activity. G.M., A.P. and J.T. designed and set up the clinical study involving sepsis and matched-control patients. A.T., C.G., P.L., V.M., F.V., P.C. and I.A.G. supervised the work, participated to the design of technical experiments and of the clinical study, and actively contributed in writing different sections of the manuscript. Y.B., S.D. and C.V. analyzed the data for the sepsis application, and Y.B., K.E., J. Baudry, A.D.G. and C.V. wrote the manuscript. All authors edited and proofread the paper.
Corresponding authors
Ethics declarations
Competing interests
Some of the authors (J. Baudry, J. Bibette, A.D.G., Y.B. & C.V.) are inventors on patent applications based on certain ideas described in this paper and may receive financial compensation via their employers’ rewards-to-inventors schemes.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Eyer, K., et al. Nat. Biotechnol. 35, 977–982 (2017): https://www.nature.com/articles/nbt.3964
Rybczynska, M., et al. 38, 5337–5342 (2020): https://doi.org/10.1016/j.vaccine.2020.05.066
Supplementary information
Supplementary Information
Supplementary Figs. 1–3 and Supplementary Method 1 (Pipeline analysis of droplet detection and tracking).
Supplementary Data 1
Complete chip design (CAD file).
Supplementary Data 2
Mask for the double-sided tape to prepare the observation chamber.
Supplementary Data 3
Mask for laser ablation.
Supplementary Data 4
Mask for the magnet holder.
Rights and permissions
About this article
Cite this article
Bounab, Y., Eyer, K., Dixneuf, S. et al. Dynamic single-cell phenotyping of immune cells using the microfluidic platform DropMap. Nat Protoc 15, 2920–2955 (2020). https://doi.org/10.1038/s41596-020-0354-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0354-0
This article is cited by
-
Harnessing 3D in vitro systems to model immune responses to solid tumours: a step towards improving and creating personalized immunotherapies
Nature Reviews Immunology (2024)
-
Single-B cell analysis correlates high-lactate secretion with stress and increased apoptosis
Scientific Reports (2024)
-
Droplet-based microfluidics
Nature Reviews Methods Primers (2023)
-
Time-resolved assessment of single-cell protein secretion by sequencing
Nature Methods (2023)
-
Pico-washing: simultaneous liquid addition and removal for continuous-flow washing of microdroplets
Microsystems & Nanoengineering (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.