Abstract
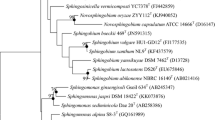
A Gram-stain-negative, aerobic, motile strain, HHU CXWT, was isolated from hair of a healthy 21-year-old female student of Hohai University, Nanjing, China. The 16S rRNA gene sequence analysis indicated that HHU CXWT represents a member of the genus Sphingomonas with the highest sequence similarity (97.6%) to the type strain S. aquatilis JSS7T. HHU CXWT grew at 4–35 °C and pH 6–8, with optimum growth at 28 °C and pH 7. Tolerance to NaCl was up to 2% (w/v) with optimum growth in 0.5–1.0% NaCl. The major fatty acids were C16:0, C17:1ω6c, C18:1ω7c11-methyl, summed feature 3 (C16:1ω7c and/or C16:1ω6c), and summed feature 8 (C18:1ω7c and/or C18:1ω6c). The predominant isoprenoid quinone was ubiquinone-10. The polar lipids were diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), sphingoglycolipid (SGL), phosphatidylinositol mannosides (PIM), and an unidentified glycolipid (GL). The DNA G + C content was 67.1%. The average nucleotide identity (ANI) values and digital DNA–DNA hybridization (dDDH) between HHU CXWT and closely related members of the genus Sphingomonas were all below the cut-off level (95–96% and 70%, respectively) for species delineation. On the basis of the phenotypic, phylogenetic and chemotaxonomic characterizations, HHU CXWT represents a novel species of the genus Sphingomonas, for which the name Sphingomonas hominis sp. nov. is proposed. The type strain is HHU CXWT (= KCTC 72946T = CGMCC 1.17504T = MCCC 1K04223T).


Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Asaf S, Numan M, Khan AL, Al-Harrasi A (2020) Sphingomonas: from diversity and genomics to functional role in environmental remediation and plant growth. Crit Rev Biotechnol 40:138–152
Aylward FO, McDonald BR, Adams SM et al (2013) Comparison of 26 sphingomonad genomes reveals diverse environmental adaptations and biodegradative capabilities. Appl Environm Microbiol 79:3724–3733
Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2, 4-diaminobutyric acid. J Appl Microbiol 48:459–470
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Cowan ST, Steel KJ (1996) Manual for the identification of medical bacteria. Cambridge University Press, London, p 232
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Feng GD, Yang SZ, Zhu HH, Li HP (2018) Emended descriptions of the species Sphingomonas adhaesiva Yabuuchi et al. 1990 and Sphingomonas ginsenosidimutans Choi et al. 2011. Int J Syst Evol Microbiol 68:970–973
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20:406–416
Galperin MY, Makarova KS, Wolf YI, Koonin EV (2015) Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–269
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Huang HY, Li J, Zhao GZ et al (2012) Sphingomonas endophytica sp. nov., isolated from Artemisia annua L. Int J Syst Evol Microbiol 62:1576–1580
Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P (2017) Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34:2115–2122
Khan AL, Waqas M, Kang S-M et al (2014) Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J Microbiol 52:689–695
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Li YQ, Rao MPN, Zhang H et al (2019) Description of Sphingomonas mesophila sp. nov., isolated from Gastrodia elata Blume. Int J Syst Evol Microbiol 69:1030–1034
Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Microbiol 47:87–95
Nurk S, Bankevich A, Antipov D et al (2013) Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737
Okonechnikov K, Golosova O, Fursov M, Team U (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004
Parte AC (2018) LPSN - List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol 68:1825–1829
Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. P Natl Acad Sci USA 106:19126–19131
Rivas R, Abril A, Trujillo ME, Velazquez E (2004) Sphingomonas phyllosphaerae sp. nov., from the phyllosphere of Acacia caven in Argentina. Int J Syst Evol Microbiol 54:2147–2150
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. Microbial ID, Inc, Newark
Siddaramappa S, Viswanathan V, Thiyagarajan S, Narjala A (2018) Genomewide characterisation of the genetic diversity of carotenogenesis in bacteria of the order Sphingomonadales. Microb Genom 4:e000172
Sievers F, Wilm A, Dineen D et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Tamaoka J (1986) Analysis of bacterial menaquinone mixtures by reverse-phase high-performance liquid chromatography. Methods Enzymol 123:251–256
Tatusova T, DiCuccio M, Badretdin A et al (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624
The Gene Ontology Consortium (2019) The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res 47:D330–D338
Yang S, Zhang X, Cao Z et al (2014) Growth-promoting Sphingomonas paucimobilis ZJSH1 associated with Dendrobium officinale through phytohormone production and nitrogen fixation. Microbial Biotechnol 7:611–620
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Acknowledgements
This research was supported by the National Natural Science Foundation of China (Nos. 31900001 and 31972856). D-F Zhang was also supported by the Fundamental Research Funds for the Central Universities (2019B02014)
Author information
Authors and Affiliations
Contributions
DFZ and ZZ designed research and project outline. XWC, DFZ, and AHZ performed isolation, deposition and polyphasic taxonomy. DFZ, and JH performed genome analysis. DFZ and AHZ drafted the manuscript. WJL revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, DF., Cui, XW., Zhao, Z. et al. Sphingomonas hominis sp. nov., isolated from hair of a 21-year-old girl. Antonie van Leeuwenhoek 113, 1523–1530 (2020). https://doi.org/10.1007/s10482-020-01460-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-020-01460-z