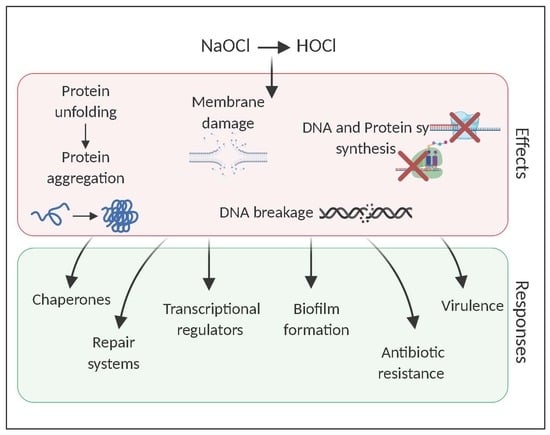
Surviving Reactive Chlorine Stress: Responses of Gram-Negative Bacteria to Hypochlorous Acid
Abstract
:1. Introduction
2. Hypochlorous Acid, a Widely Used Disinfectant
2.1. The Potent Antimicrobial Effects of HOCl
2.1.1. Reaction of HOCl with Sulfur-Containing Amino Acids
2.1.2. Reaction of HOCl with Aromatic Amino Acids
2.1.3. Reaction of HOCl with Nitrogen-Containing Compounds
2.1.4. Reaction of HOCl with Lipids
3. Adaptive Response of Gram-Negative Cells to HOCl
3.1. Chaperones: The Immediate Response against HOCl-Induced Stress
3.2. Transcriptional Regulators
3.3. Formation of Biofilms as an Adaptive Response to HOCl
3.4. Sublethal Concentrations of HOCl Induce Antimicrobial Resistance and Virulence Gene Expression and VBNC
4. Final Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Morris, S.; Cerceo, E. Trends, Epidemiology, and Management of Multi-Drug Resistant Gram-Negative Bacterial Infections in the Hospitalized Setting. Antibiotics 2020, 9, 196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Cleaning and Disinfection of Environmental Surfaces in the Context of COVID-19: Interim Guidance; World Health Organization: Geneva, Switzerland, 2020; Available online: https://www.who.int/publications/i/item/cleaning-and-disinfection-of-environmental-surfaces-inthe-context-of-covid-19 (accessed on 10 August 2020).
- Koebnik, R.; Locher, K.P.; Gelder, P.V. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol. Microbiol. 2000, 37, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. Multidrug resistance in Gram-negative bacteria. Curr. Opin. Microbiol. 2001, 4, 500–508. [Google Scholar] [CrossRef]
- Cox, G.; Wright, G.D. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef]
- Blair, J.M.; Richmond, G.E.; Piddock, L.J. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014, 9, 1165–1177. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [Green Version]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Crouzet, M.; Le Senechal, C.; Brözel, V.S.; Costaglioli, P.; Barthe, C.; Bonneu, M.; Garbay, B.; Vilain, S. Exploring early steps in biofilm formation: Set-up of an experimental system for molecular studies. BMC Microbiol. 2014, 14, 253–264. [Google Scholar] [CrossRef] [Green Version]
- Whiteley, M.; Bangera, M.G.; Bumgarner, R.E.; Parsek, M.R.; Teitzel, G.M.; Lory, S.; Greenberg, E.P. Gene expression in Pseudomonas aeruginosa bio®lms. Bind. Protein 2001, 413, 860–864. [Google Scholar]
- Sauer, K. The genomics and proteomics of biofilm formation. Genome Biol. 2003, 4, 219. [Google Scholar] [CrossRef] [Green Version]
- Hall-Stoodley, L.; Stoodley, P. Evolving concepts in biofilm infections. Cell. Microbiol. 2009, 11, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Pullanhi, U.; Khan, S.; Vinod, V.; Mohan, K.; Kumar, A. Outcome of Acute Urinary Tract Infections Caused by Uropathogenic Escherichia coli with Phenotypically Demonstrable Virulence Factors. Ann. Afr. Med. 2019, 18, 138–142. [Google Scholar] [PubMed]
- Zalewska-Piątek, B.M.; Piątek, R.J. Alternative treatment approaches of urinary tract infections caused by uropathogenic Escherichia coli strains—Review. Acta Biochim. Pol. 2019, 66, 129–138. [Google Scholar] [PubMed]
- Speranza, B.; Corbo, M.R. Chapter 11—The Impact of Biofilms on Food Spoilage. In The Microbiological Quality of Food; Woodhead Publishing Series in Food Science, Technology and Nutrition; Bevilacqua, A., Corbo, M.R., Sinigaglia, M., Eds.; Woodhead Publishing: Cambridge, UK, 2017; pp. 259–282. ISBN 978-0-08-100502-6. [Google Scholar]
- Sofos, J.N.; Geornaras, I. Overview of current meat hygiene and safety risks and summary of recent studies on biofilms, and control of Escherichia coli O157:H7 in nonintact, and Listeria monocytogenes in ready-to-eat, meat products. Meat Sci. 2010, 86, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Bassler, B.L. Surviving as a Community: Antibiotic Tolerance and Persistence in Bacterial Biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef]
- Qureshi, N. 18-Beneficial biofilms: Wastewater and other industrial applications. In Biofilms in the Food and Beverage Industries; Woodhead Publishing Series in Food Science, Technology and Nutrition; Fratamico, P.M., Annous, B.A., Gunther, N.W., Eds.; Woodhead Publishing: Cambridge, UK, 2009; pp. 474–498. ISBN 978-1-84569-477-7. [Google Scholar]
- Lear, G. Biofilms in Bioremediation: Current Research and Emerging Technologies; Caister Academic Press: London, UK, 2016; ISBN 978-1-910190-29-6. [Google Scholar]
- Robertson, S.R.; McLean, R.J.C. Department of Biology, Texas State University, 601 University Drive, San Marcos, TX 78666, USA Beneficial biofilms. AIMS Bioeng. 2015, 2, 437–448. [Google Scholar] [CrossRef]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [Green Version]
- Anderl, J.N.; Zahller, J.; Roe, F.; Stewart, P.S. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 2003, 47, 1251–1256. [Google Scholar] [CrossRef] [Green Version]
- Walters, M.C.; Roe, F.; Bugnicourt, A.; Franklin, M.J.; Stewart, P.S. Contributions of Antibiotic Penetration, Oxygen Limitation, and Low Metabolic Activity to Tolerance of Pseudomonas aeruginosa Biofilms to Ciprofloxacin and Tobramycin. Antimicrob. Agents Chemother. 2003, 47, 317–323. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Bryers, J.D. Non-invasive determination of conjugative transfer of plasmids bearing antibiotic-resistance genes in biofilm-bound bacteria: Effects of substrate loading and antibiotic selection. Appl. Microbiol. Biotechnol. 2013, 97, 317–328. [Google Scholar] [CrossRef] [Green Version]
- Nazir, R.; Zaffar, M.R.; Amin, I. Chapter 8—Bacterial biofilms: The remarkable heterogeneous biological communities and nitrogen fixing microorganisms in lakes. In Freshwater Microbiology; Bandh, S.A., Shafi, S., Shameem, N., Eds.; Academic Press: London, UK, 2019; pp. 307–340. ISBN 978-0-12-817495-1. [Google Scholar]
- Frieri, M.; Kumar, K.; Boutin, A. Antibiotic resistance. J. Infect. Public Health 2017, 10, 369–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, V.E.; Bushnell, D.; Passador, L.; Brooks, A.I.; Iglewski, B.H. Microarray Analysis of Pseudomonas aeruginosa Quorum-Sensing Regulons: Effects of Growth Phase and Environment. J. Bacteriol. 2003, 185, 2080–2095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fila, G.; Krychowiak, M.; Rychlowski, M.; Bielawski, K.P.; Grinholc, M. Antimicrobial blue light photoinactivation of Pseudomonas aeruginosa: Quorum sensing signaling molecules, biofilm formation and pathogenicity. J. Biophotonics 2018, 11, e201800079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.; Kong, J.; Dong, B.; Huang, H.; Wang, K.; Wu, L.; Hou, C.; Liang, Y.; Li, B.; Chen, Y. Baicalein attenuates the quorum sensing-controlled virulence factors of Pseudomonas aeruginosa and relieves the inflammatory response in P. aeruginosa-infected macrophages by downregulating the MAPK and NFκB signal-transduction pathways. Drug Des. Devel. Ther. 2016, 10, 183–203. [Google Scholar] [CrossRef] [Green Version]
- Tielen, P.; Rosenau, F.; Wilhelm, S.; Jaeger, K.-E.; Flemming, H.-C.; Wingender, J. Extracellular enzymes affect biofilm formation of mucoid Pseudomonas aeruginosa. Microbiology 2010, 156, 2239–2252. [Google Scholar] [CrossRef] [Green Version]
- White, G.C. White’s Handbook of Chlorination and Alternative Disinfectants; Wiley: Hoboken, NJ, USA, 2010; ISBN 978-0-470-18098-3. [Google Scholar]
- Fukuzaki, S. Mechanisms of Actions of Sodium Hypochlorite in Cleaning and Disinfection Processes. Biocontrol. Sci. 2006, 11, 147–157. [Google Scholar] [CrossRef]
- Rutala, W.A.; Weber, D.J. Uses of inorganic hypochlorite (bleach) in health-care facilities. Clin. Microbiol. Rev. 1997, 10, 597–610. [Google Scholar] [CrossRef]
- Zwiener, C.; Richardson, S.D.; De Marini, D.M.; Grummt, T.; Glauner, T.; Frimmel, F.H. Drowning in Disinfection Byproducts? Assessing Swimming Pool Water. Environ. Sci. Technol. 2007, 41, 363–372. [Google Scholar] [CrossRef]
- Deborde, M.; Von Gunten, U. Reactions of chlorine with inorganic and organic compounds during water treatment—kinetics and mechanisms: A critical review. Water Res. 2008, 42, 13–51. [Google Scholar]
- Wahman, D.G.; Pressman, J.G. Distribution System Residuals—Is “Detectable” Still Acceptable for Chloramines? J. AWWA 2015, 107, 53–63. [Google Scholar] [CrossRef]
- Health Canada. Guidelines for Canadian Drinking Water Quality: Guideline Technical Document—Chlorine; Health Canada: Ottawa, ON, Canada, 2009.
- Patel, P.; Sanghvi, S.; Malik, K.; Khachemoune, A. Back to the basics: Diluted bleach for COVID-19. J. Am. Acad. Dermatol. 2020, 83, 279–280. [Google Scholar] [CrossRef] [PubMed]
- Marchitelli, R. Canadians are Accidentally Poisoning Themselves while Cleaning to Prevent COVID-19|CBC News. Available online: https://www.cbc.ca/news/health/covid-19-accidental-poisoning-cleaning-products-1.5552779 (accessed on 29 June 2020).
- Lin, Q.; Lim, J.Y.C.; Xue, K.; Yew, P.Y.M.; Owh, C.; Chee, P.L.; Loh, X.J. Sanitizing agents for virus inactivation and disinfection. View 2020, 1, e16–e41. [Google Scholar] [CrossRef]
- Campagna, M.V.; Faure-Kumar, E.; Treger, J.A.; Cushman, J.D.; Grogan, T.R.; Kasahara, N.; Lawson, G.W. Factors in the Selection of Surface Disinfectants for Use in a Laboratory Animal Setting. J. Am. Assoc. Lab. Anim. Sci. JAALAS 2016, 55, 175–188. [Google Scholar] [PubMed]
- Slaughter, R.J.; Watts, M.; Vale, J.A.; Grieve, J.R.; Schep, L.J. The clinical toxicology of sodium hypochlorite. Clin. Toxicol. 2019, 57, 303–311. [Google Scholar] [CrossRef]
- Wang, L.; Bassiri, M.; Najafi, R.; Najafi, K.; Yang, J.; Khosrovi, B.; Hwong, W.; Barati, E.; Belisle, B.; Celeri, C.; et al. Hypochlorous Acid as a Potential Wound Care Agent. J. Burns Wounds 2007, 6, 65–79. [Google Scholar]
- Robson, M.C.; Payne, W.G.; Ko, F.; Mentis, M.; Donati, G.; Shafii, S.M.; Culverhouse, S.; Wang, L.; Khosrovi, B.; Najafi, R.; et al. Hypochlorous Acid as a Potential Wound Care Agent. J. Burns Wounds 2007, 6, 80–90. [Google Scholar]
- Gold, M.H.; Andriessen, A.; Bhatia, A.C.; Bitter, P.; Chilukuri, S.; Cohen, J.L.; Robb, C.W. Topical stabilized hypochlorous acid: The future gold standard for wound care and scar management in dermatologic and plastic surgery procedures. J. Cosmet. Dermatol. 2020, 19, 270–277. [Google Scholar] [CrossRef]
- Kettle, A.J.; Winterbourn, C.C. Myeloperoxidase: A key regulator of neutrophil oxidant production. Redox Rep. 1997, 3, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Winterbourn, C.C. Biological reactivity and biomarkers of the neutrophil oxidant, hypochlorous acid. Toxicology 2002, 181–182, 223–227. [Google Scholar] [CrossRef]
- Mütze, S.; Hebling, U.; Stremmel, W.; Wang, J.; Arnhold, J.; Pantopoulos, K.; Mueller, S. Myeloperoxidase-derived Hypochlorous Acid Antagonizes the Oxidative Stress-mediated Activation of Iron Regulatory Protein 1. J. Biol. Chem. 2003, 278, 40542–40549. [Google Scholar] [CrossRef] [Green Version]
- Weiss, S.J. Tissue Destruction by Neutrophils. N. Engl. J. Med. 1989, 320, 365–376. [Google Scholar] [PubMed]
- Winter, J.; Ilbert, M.; Graf, P.C.F.; Özcelik, D.; Jakob, U. Bleach Activates a Redox-Regulated Chaperone by Oxidative Protein Unfolding. Cell 2008, 135, 691–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrette, W.; Albrich, J.; Hurst, J. Hypochlorous acid-promoted loss of metabolic energy in Escherichia coli. Infect. Immun. 1987, 55, 2518–2525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrette, W.C., Jr.; Hannum, D.M.; Wheeler, W.D.; Hurst, J.K. General mechanism for the bacterial toxicity of hypochlorous acid: Abolition of ATP production. Biochemistry 1989, 28, 9172–9178. [Google Scholar] [CrossRef]
- Dukan, S.; Touati, D. Hypochlorous acid stress in Escherichia coli: Resistance, DNA damage, and comparison with hydrogen peroxide stress. J. Bacteriol. 1996, 178, 6145–6150. [Google Scholar] [CrossRef] [Green Version]
- Albrich, J.M.; Gilbaugh, J.H.; Callahan, K.B.; Hurst, J.K. Effects of the putative neutrophil-generated toxin, hypochlorous acid, on membrane permeability and transport systems of Escherichia coli. J. Clin. Invest. 1986, 78, 177–184. [Google Scholar] [CrossRef] [Green Version]
- McKenna, S.M.; Davies, K.J.A. The inhibition of bacterial growth by hypochlorous acid. Possible role in the bactericidal activity of phagocytes. Biochem. J. 1988, 254, 685–692. [Google Scholar] [CrossRef] [Green Version]
- Rosen, H.; Klebanoff, S.J.; Wang, Y.; Brot, N.; Heinecke, J.W.; Fu, X. Methionine oxidation contributes to bacterial killing by the myeloperoxidase system of neutrophils. Proc. Natl. Acad. Sci. USA 2009, 106, 18686–18691. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, C.L. Hypochlorous acid-mediated modification of proteins and its consequences. Essays Biochem. 2020, 64, 75–86. [Google Scholar] [CrossRef]
- Hawkins, C.L.; Pattison, D.I.; Davies, M.J. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids 2003, 25, 259–274. [Google Scholar] [CrossRef]
- Kiamco, M.M.; Zmuda, H.M.; Mohamed, A.; Call, D.R.; Raval, Y.S.; Patel, R.; Beyenal, H. Hypochlorous-Acid-Generating Electrochemical Scaffold for Treatment of Wound Biofilms. Sci. Rep. 2019, 9, 2683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkins, C.L.; Davies, M.J. Hypochlorite-Induced Damage to DNA, RNA, and Polynucleotides: Formation of Chloramines and Nitrogen-Centered Radicals. Chem. Res. Toxicol. 2002, 15, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Dukan, S.; Belkin, S.; Touati, D. Reactive Oxygen Species Are Partially Involved in the Bacteriocidal Action of Hypochlorous Acid. Arch. Biochem. Biophys. 1999, 367, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Rosen, H.; Klebanoff, S.J. Oxidation of Escherichia coli iron centers by the myeloperoxidase-mediated microbicidal system. J. Biol. Chem. 1982, 257, 13731–13735. [Google Scholar]
- Candeias, L.P.; Stratford, M.R.; Wardman, P. Formation of hydroxyl radicals on reaction of hypochlorous acid with ferrocyanide, a model iron (II) complex. Free Radic. Res. 1994, 20, 241–249. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Comparative reactivities of various biological compounds with myeloperoxidase-hydrogen peroxide-chloride, and similarity of oxidant to hypochlorite. Biochim. Biophys. Acta BBA Gen. Subj. 1985, 840, 204–210. [Google Scholar] [CrossRef]
- Van Bergen, L.A.H.; Roos, G.; De Proft, F. From Thiol to Sulfonic Acid: Modeling the Oxidation Pathway of Protein Thiols by Hydrogen Peroxide. J. Phys. Chem. A 2014, 118, 6078–6084. [Google Scholar] [CrossRef]
- Fu, X.; Mueller, D.M.; Heinecke, J.W. Generation of Intramolecular and Intermolecular Sulfenamides, Sulfinamides, and Sulfonamides by Hypochlorous Acid: A Potential Pathway for Oxidative Cross-Linking of Low-Density Lipoprotein by Myeloperoxidase. Biochemistry 2002, 41, 1293–1301. [Google Scholar]
- Pattison, D.I.; Davies, M.J.; Hawkins, C.L. Reactions and reactivity of myeloperoxidase-derived oxidants: Differential biological effects of hypochlorous and hypothiocyanous acids. Free Radic. Res. 2012, 46, 975–995. [Google Scholar] [CrossRef]
- Nagy, P.; Ashby, M.T. Reactive Sulfur Species: Kinetics and Mechanism of the Oxidation of Cystine by Hypochlorous Acid to Give N,N‘-Dichlorocystine. Chem. Res. Toxicol. 2005, 18, 919–923. [Google Scholar] [CrossRef]
- Curtis, M.P.; Hicks, A.J.; Neidigh, J.W. Kinetics of 3-chlorotyrosine formation and loss due to hypochlorous acid and chloramines. Chem. Res. Toxicol. 2011, 24, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.J.; Wholey, W.-Y.; Jakob, U. Bacterial Responses to Reactive Chlorine Species. Annu. Rev. Microbiol. 2013, 67, 141–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, A.L.; Winterbourn, C.C.; Brennan, S.O.; Jordan, T.W.; Kettle, A.J. Characterization of non-covalent oligomers of proteins treated with hypochlorous acid. Biochem. J. 2003, 375, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Spickett, C.M.; Jerlich, A.; Panasenko, O.M.; Arnhold, J.; Pitt, A.R.; Stelmaszyñska, T.; Schaur, J. The reactions of hypochlorous acid, the reactive oxygen species produced by myeloperoxidase, with lipids. Acta Biochim. Pol. 2000, 47, 889–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dever, G.; Wainwright, C.L.; Kennedy, S.; Spickett, C.M. Fatty acid and phospholipid chlorohydrins cause cell stress and endothelial adhesion. Acta Biochim. Pol. 2006, 53, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Niki, E. Lipid peroxidation: Physiological levels and dual biological effects. Free Radic. Biol. Med. 2009, 47, 469–484. [Google Scholar] [CrossRef]
- Small, D.A.; Chang, W.; Toghrol, F.; Bentley, W.E. Comparative global transcription analysis of sodium hypochlorite, peracetic acid, and hydrogen peroxide on Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2007, 76, 1093–1105. [Google Scholar] [CrossRef]
- Bodet, C.; Sahr, T.; Dupuy, M.; Buchrieser, C.; Héchard, Y. Legionella pneumophila transcriptional response to chlorine treatment. Water Res. 2012, 46, 808–816. [Google Scholar]
- Wang, S.; Deng, K.; Zaremba, S.; Deng, X.; Lin, C.; Wang, Q.; Tortorello, M.L.; Zhang, W. Transcriptomic Response of Escherichia coli O157:H7 to Oxidative Stress. Appl. Environ. Microbiol. 2009, 75, 6110–6123. [Google Scholar] [CrossRef] [Green Version]
- Goemans, C.V.; Collet, J.-F. Stress-induced chaperones: A first line of defense against the powerful oxidant hypochlorous acid. F1000Research 2019, 8, 1678–1684. [Google Scholar] [CrossRef]
- Groitl, B.; Dahl, J.-U.; Schroeder, J.W.; Jakob, U. Pseudomonas aeruginosa defense systems against microbicidal oxidants. Mol. Microbiol. 2017, 106, 335–350. [Google Scholar] [CrossRef] [Green Version]
- Small, D.A.; Chang, W.; Toghrol, F.; Bentley, W.E. Toxicogenomic analysis of sodium hypochlorite antimicrobial mechanisms in Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2007, 74, 176–185. [Google Scholar] [CrossRef]
- Melnyk, R.A.; Youngblut, M.D.; Clark, I.C.; Carlson, H.K.; Wetmore, K.M.; Price, M.N.; Iavarone, A.T.; Deutschbauer, A.M.; Arkin, A.P.; Coates, J.D. Novel Mechanism for Scavenging of Hypochlorite Involving a Periplasmic Methionine-Rich Peptide and Methionine Sulfoxide Reductase. mBio 2015, 6, e00233-15. [Google Scholar] [CrossRef] [Green Version]
- Morales, E.H.; Calderón, I.L.; Collao, B.; Gil, F.; Porwollik, S.; McClelland, M.; Saavedra, C.P. Hypochlorous acid and hydrogen peroxide-induced negative regulation of Salmonella enterica serovar Typhimurium ompW by the response regulator ArcA. BMC Microbiol. 2012, 12, 63–73. [Google Scholar] [CrossRef] [Green Version]
- Salazar, J.K.; Deng, K.; Tortorello, M.L.; Brandl, M.T.; Wang, H.; Zhang, W. Genes ycfR, sirA and yigG Contribute to the Surface Attachment of Salmonella enterica Typhimurium and Saintpaul to Fresh Produce. PLoS ONE 2013, 8, e57272. [Google Scholar] [CrossRef]
- Gambino, M.; Cappitelli, F. Full article: Mini-review: Biofilm responses to oxidative stress. Biofouling 2016, 32, 167–178. [Google Scholar] [CrossRef]
- Sultana, S.; Foti, A.; Dahl, J.-U. Bacterial Defense Systems against the Neutrophilic Oxidant Hypochlorous Acid. Infect. Immun. 2020. [Google Scholar] [CrossRef]
- Voth, W.; Jakob, U. Stress-Activated Chaperones: A First Line of Defense. Trends Biochem. Sci. 2017, 42, 899–913. [Google Scholar] [CrossRef] [PubMed]
- Pattison, D.I.; Davies, M.J. Absolute Rate Constants for the Reaction of Hypochlorous Acid with Protein Side Chains and Peptide Bonds. Chem. Res. Toxicol. 2001, 14, 1453–1464. [Google Scholar] [CrossRef] [PubMed]
- Buchner, J. Molecular chaperones and protein quality control: An introduction to the JBC Reviews thematic series. J. Biol. Chem. 2019, 294, 2074–2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef]
- Wholey, W.Y.; Jakob, U. Hsp33 confers bleach resistance by protecting elongation factor Tu against oxidative degradation in Vibrio cholerae. Mol. Microbiol. 2012, 83, 981–991. [Google Scholar] [CrossRef] [Green Version]
- Jakob, U.; Eser, M.; Bardwell, J.C.A. Redox Switch of Hsp33 Has a Novel Zinc-binding Motif. J. Biol. Chem. 2000, 275, 38302–38310. [Google Scholar] [CrossRef] [Green Version]
- Krewing, M.; Stepanek, J.J.; Cremers, C.; Lackmann, J.-W.; Schubert, B.; Müller, A.; Awakowicz, P.; Leichert, L.I.O.; Jakob, U.; Bandow, J.E. The molecular chaperone Hsp33 is activated by atmospheric-pressure plasma protecting proteins from aggregation. J. R. Soc. Interface 2019, 16, 20180966. [Google Scholar] [CrossRef]
- Hoffmann, J.H.; Linke, K.; Graf, P.C.; Lilie, H.; Jakob, U. Identification of a redox-regulated chaperone network. EMBO J. 2004, 23, 160–168. [Google Scholar] [CrossRef] [Green Version]
- Bruel, N.; Castanié-Cornet, M.-P.; Cirinesi, A.-M.; Koningstein, G.; Georgopoulos, C.; Luirink, J.; Genevaux, P. Hsp33 controls elongation factor-Tu stability and allows Escherichia coli growth in the absence of the major DnaK and trigger factor chaperones. J. Biol. Chem. 2012, 287, 44435–44446. [Google Scholar] [CrossRef] [Green Version]
- Jakob, U.; Muse, W.; Eser, M.; Bardwell, J.C.A. Chaperone Activity with a Redox Switch. Cell 1999, 96, 341–352. [Google Scholar] [CrossRef] [Green Version]
- Winter, J.; Linke, K.; Jatzek, A.; Jakob, U. Severe Oxidative Stress Causes Inactivation of DnaK and Activation of the Redox-Regulated Chaperone Hsp33. Mol. Cell 2005, 17, 381–392. [Google Scholar] [CrossRef]
- Ilbert, M.; Horst, J.; Ahrens, S.; Winter, J.; Graf, P.C.F.; Lilie, H.; Jakob, U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat. Struct. Mol. Biol. 2007, 14, 556–563. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Kwon, A.-R.; Lee, B.-J. A novel chlorination-induced ribonuclease YabJ from Staphylococcus aureus. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [Green Version]
- Müller, A.; Langklotz, S.; Lupilova, N.; Kuhlmann, K.; Bandow, J.E.; Leichert, L.I.O. Activation of RidA chaperone function by N -chlorination. Nat. Commun. 2014, 5, 5804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goemans, C.V.; Beaufay, F.; Arts, I.S.; Agrebi, R.; Vertommen, D.; Collet, J.-F. The Chaperone and Redox Properties of CnoX Chaperedoxins Are Tailored to the Proteostatic Needs of Bacterial Species. mBio 2018, 9, e01541-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goemans, C.V.; Vertommen, D.; Agrebi, R.; Collet, J.-F. CnoX Is a Chaperedoxin: A Holdase that Protects Its Substrates from Irreversible Oxidation. Mol. Cell 2018, 70, 614.e7–627.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Abreu Meireles, D.; Yokomizo, C.; Netto, L. Investigation on the requirements for YbbN/CnoX displaying thiol-disulfide oxidoreductase and chaperone activities. Eur. PMC 2020. [Google Scholar] [CrossRef] [Green Version]
- Gray, M.J.; Wholey, W.-Y.; Wagner, N.O.; Cremers, C.M.; Mueller-Schickert, A.; Hock, N.T.; Krieger, A.G.; Smith, E.M.; Bender, R.A.; Bardwell, J.C.A.; et al. Polyphosphate Is a Primordial Chaperone. Mol. Cell 2014, 53, 689–699. [Google Scholar] [CrossRef] [Green Version]
- Gray, M.J.; Jakob, U. Oxidative stress protection by polyphosphate—New roles for an old player. Curr. Opin. Microbiol. 2015, 24, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Gray, M.J. Interactions between DksA and Stress-Responsive Alternative Sigma Factors Control Inorganic Polyphosphate Accumulation in Escherichia coli. J. Bacteriol. 2020, 202. [Google Scholar] [CrossRef]
- Ferguson, G.P.; Tötemeyer, S.; MacLean, M.J.; Booth, I.R. Methylglyoxal production in bacteria: Suicide or survival? Arch. Microbiol. 1998, 170, 209–218. [Google Scholar] [CrossRef]
- Browning, D.F.; Busby, S.J. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004, 2, 57–65. [Google Scholar] [CrossRef]
- Parker, B.W.; Schwessinger, E.A.; Jakob, U.; Gray, M.J. The RclR Protein Is a Reactive Chlorine-specific Transcription Factor in Escherichia coli. J. Biol. Chem. 2013, 288, 32574–32584. [Google Scholar] [CrossRef] [Green Version]
- Baek, Y.; Kim, J.; Ahn, J.; Jo, I.; Hong, S.; Ryu, S.; Ha, N.-C. Structure and function of the hypochlorous acid–induced flavoprotein RclA from Escherichia coli. J. Biol. Chem. 2020, 295, 3202–3212. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.J.; Wholey, W.-Y.; Parker, B.W.; Kim, M.; Jakob, U. NemR Is a Bleach-sensing Transcription Factor. J. Biol. Chem. 2013, 288, 13789–13798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebendorfer, K.M.; Drazic, A.; Le, Y.; Gundlach, J.; Bepperling, A.; Kastenmüller, A.; Ganzinger, K.A.; Braun, N.; Franzmann, T.M.; Winter, J. Identification of a Hypochlorite-specific Transcription Factor from Escherichia coli. J. Biol. Chem. 2012, 287, 6892–6903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.; Do, E.; Kim, M.; Park, H.-J.; Lee, J.; Han, S.-W. A LysR-Type Transcriptional Regulator LcrX Is Involved in Virulence, Biofilm Formation, Swimming Motility, Siderophore Secretion, and Growth in Sugar Sources in Xanthomonas axonopodis Pv. glycines. Front. Plant Sci. 2020, 10, 1657–1675. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, B.; Yang, P.; Wang, T.; Chang, Z.; Wang, J.; Wang, Q.; Li, W.; Wu, J.; Huang, D.; et al. LysR-type transcriptional regulator OvrB encoded in O island 9 drives enterohemorrhagic Escherichia coli O157:H7 virulence. Virulence 2019, 10, 783–792. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Kim, S.H.; Ahn, J.; Jo, I.; Lee, Z.-W.; Choi, S.H.; Ha, N.-C. Crystal Structure of the Regulatory Domain of MexT, a Transcriptional Activator of the MexEF-OprN Efflux Pump in Pseudomonas aeruginosa. Mol. Cells 2019, 42, 850–857. [Google Scholar]
- Fu, Y.; Cai, Q.; Wang, Y.; Li, W.; Yu, J.; Yang, G.; Lin, W.; Lin, X. Four LysR-type transcriptional regulator family proteins (LTTRs) involved in antibiotic resistance in Aeromonas hydrophila. World J. Microbiol. Biotechnol. 2019, 35, 127. [Google Scholar] [CrossRef]
- Santiago, A.S.; Santos, C.A.; Mendes, J.S.; Toledo, M.A.S.; Beloti, L.L.; Souza, A.A.; Souza, A.P. Characterization of the LysR-type transcriptional regulator YcjZ-like from Xylella fastidiosa overexpressed in Escherichia coli. Protein Expr. Purif. 2015, 113, 72–78. [Google Scholar] [CrossRef] [Green Version]
- Reen, F.J.; Haynes, J.M.; Mooij, M.J.; O’Gara, F. A Non-Classical LysR-Type Transcriptional Regulator PA2206 Is Required for an Effective Oxidative Stress Response in Pseudomonas aeruginosa. PLoS ONE 2013, 8, e54479. [Google Scholar] [CrossRef] [Green Version]
- Jo, I.; Kim, D.; No, T.; Hong, S.; Ahn, J.; Ryu, S.; Ha, N.-C. Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator. Proc. Natl. Acad. Sci. USA 2019, 116, 3740–3745. [Google Scholar] [CrossRef] [Green Version]
- Cornelis, P.; Wei, Q.; Andrews, S.C.; Vinckx, T. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 2011, 3, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Drazic, A.; Miura, H.; Peschek, J.; Le, Y.; Bach, N.C.; Kriehuber, T.; Winter, J. Methionine oxidation activates a transcription factor in response to oxidative stress. Proc. Natl. Acad. Sci. USA 2013, 110, 9493–9498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drazic, A.; Tsoutsoulopoulos, A.; Peschek, J.; Gundlach, J.; Krause, M.; Bach, N.C.; Gebendorfer, K.M.; Winter, J. Role of cysteines in the stability and DNA-binding activity of the hypochlorite-specific transcription factor HypT. PLoS ONE 2013, 8, e75683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drazic, A.; Gebendorfer, K.M.; Mak, S.; Steiner, A.; Krause, M.; Bepperling, A.; Winter, J. Tetramers are the activation-competent species of the HOCl-specific transcription factor HypT. J. Biol. Chem. 2014, 289, 977–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makukhin, N.; Havelka, V.; Poláchová, E.; Rampírová, P.; Tarallo, V.; Strisovsky, K.; Míšek, J. Resolving oxidative damage to methionine by an unexpected membrane-associated stereoselective reductase discovered using chiral fluorescent probes. FEBS J. 2019, 286, 4024–4035. [Google Scholar] [CrossRef]
- Feige, M.J. Oxidative Folding of Proteins: Basic Principles, Cellular Regulation and Engineering; Royal Society of Chemistry: London, UK, 2018; ISBN 978-1-78262-990-0. [Google Scholar]
- Nagl, M.; Arnitz, R.; Lackner, M. N-Chlorotaurine, a Promising Future Candidate for Topical Therapy of Fungal Infections. Mycopathologia 2018, 183, 161–170. [Google Scholar] [CrossRef] [Green Version]
- Derke, R.M.; Barron, A.J.; Billiot, C.E.; Chaple, I.F.; Lapi, S.E.; Broderick, N.A.; Gray, M.J. The Cu (II) reductase RclA protects Escherichia coli against the combination of hypochlorous acid and intracellular copper. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Shin, J.; Park, C. Novel regulatory system nemRA–gloA for electrophile reduction in Escherichia coli K-12. Mol. Microbiol. 2013, 88, 395–412. [Google Scholar] [CrossRef]
- Ozyamak, E.; de Almeida, C.; de Moura, A.P.S.; Miller, S.; Booth, I.R. Integrated stress response of Escherichia coli to methylglyoxal: Transcriptional readthrough from the nemRA operon enhances protection through increased expression of glyoxalase I. Mol. Microbiol. 2013, 88, 936–950. [Google Scholar] [CrossRef] [Green Version]
- Fitzpatrick, T.B.; Amrhein, N.; Macheroux, P. Characterization of YqjM, an Old Yellow Enzyme Homolog from Bacillus subtilis Involved in the Oxidative Stress Response. J. Biol. Chem. 2003, 278, 19891–19897. [Google Scholar] [CrossRef] [Green Version]
- Gray, M.J.; Li, Y.; Leichert, L.I.-O.; Xu, Z.; Jakob, U. Does the Transcription Factor NemR Use a Regulatory Sulfenamide Bond to Sense Bleach? Antioxid. Redox Signal. 2015, 23, 747–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subhadra, B.; Surendran, S.; Kim, D.H.; Woo, K.; Oh, M.H.; Choi, C.H. The transcription factor NemR is an electrophile-sensing regulator important for the detoxification of reactive electrophiles in Acinetobacter nosocomialis. Res. Microbiol. 2019, 170, 123–130. [Google Scholar] [CrossRef]
- Hillion, M.; Antelmann, H. Thiol-based redox switches in prokaryotes. Biol. Chem. 2015, 396, 415–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubbs, J.M.; Mongkolsuk, S. Peroxide-sensing transcriptional regulators in bacteria. J. Bacteriol. 2012, 194, 5495–5503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charoenlap, N.; Sornchuer, P.; Piwkam, A.; Srijaruskul, K.; Mongkolsuk, S.; Vattanaviboon, P. The roles of peroxide protective regulons in protecting Xanthomonas campestris pv. campestris from sodium hypochlorite stress. Can. J. Microbiol. 2015, 61, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Phillippy, A.M.; Deng, K.; Rui, X.; Li, Z.; Tortorello, M.L.; Zhang, W. Transcriptomic Responses of Salmonella enterica Serovars Enteritidis and Typhimurium to Chlorine-Based Oxidative Stress. Appl. Environ. Microbiol. 2010, 76, 5013–5024. [Google Scholar] [CrossRef] [Green Version]
- Maddocks, S.E.; Oyston, P.C.F. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008, 154, 3609–3623. [Google Scholar] [CrossRef] [Green Version]
- Giró, M.; Carrillo, N.; Krapp, A.R. Glucose-6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli. Microbiology 2006, 152, 1119–1128. [Google Scholar] [CrossRef] [Green Version]
- Semchyshyn, H.; Bagnyukova, T.; Storey, K.; Lushchak, V. Hydrogen peroxide increases the activities of soxRS regulon enzymes and the levels of oxidized proteins and lipids in Escherichia coli. Cell Biol. Int. 2005, 29, 898–902. [Google Scholar] [CrossRef]
- Zheng, M.; Doan, B.; Schneider, T.D.; Storz, G. OxyR and SoxRS Regulation offur. J. Bacteriol. 1999, 181, 4639–4643. [Google Scholar] [CrossRef] [Green Version]
- Michán, C.; Manchado, M.; Pueyo, C. SoxRS Down-Regulation of rob Transcription. J. Bacteriol. 2002, 184, 4733–4738. [Google Scholar] [CrossRef] [Green Version]
- Dukan, S.; Dadon, S.; Smulski, D.R.; Belkin, S. Hypochlorous acid activates the heat shock and soxRS systems of Escherichia coli. Appl. Environ. Microbiol. 1996, 62, 4003–4008. [Google Scholar] [CrossRef] [Green Version]
- Collao, B.; Morales, E.; Gil, F.; Polanco, R.; Calderón, I.; Saavedra, C. Differential expression of the transcription factors MarA, Rob, and SoxS of Salmonella Typhimurium in response to sodium hypochlorite: Down-regulation of rob by MarA and SoxS. Arch. Microbiol. 2012, 194, 933–942. [Google Scholar] [CrossRef]
- Pardo-Esté, C.; Castro-Severyn, J.; Krüger, G.I.; Cabezas, C.E.; Briones, A.C.; Aguirre, C.; Morales, N.; Baquedano, M.S.; Sulbaran, Y.N.; Hidalgo, A.A.; et al. The Transcription Factor ArcA Modulates Salmonella’s Metabolism in Response to Neutrophil Hypochlorous Acid-Mediated Stress. Front. Microbiol. 2019, 10, 2754–2765. [Google Scholar] [CrossRef] [Green Version]
- Pardo-Esté, C.; Hidalgo, A.A.; Aguirre, C.; Briones, A.C.; Cabezas, C.E.; Castro-Severyn, J.; Fuentes, J.A.; Opazo, C.M.; Riedel, C.A.; Otero, C.; et al. The ArcAB two-component regulatory system promotes resistance to reactive oxygen species and systemic infection by Salmonella Typhimurium. PLoS ONE 2018, 13, e0203497. [Google Scholar] [CrossRef] [Green Version]
- Cabezas, C.E.; Briones, A.C.; Aguirre, C.; Pardo-Esté, C.; Castro-Severyn, J.; Salinas, C.R.; Baquedano, M.S.; Hidalgo, A.A.; Fuentes, J.A.; Morales, E.H.; et al. The transcription factor SlyA from Salmonella Typhimurium regulates genes in response to hydrogen peroxide and sodium hypochlorite. Res. Microbiol. 2018, 169, 263–278. [Google Scholar] [CrossRef]
- Landini, P. Cross-talk mechanisms in biofilm formation and responses to environmental and physiological stress in Escherichia coli. Res. Microbiol. 2009, 160, 259–266. [Google Scholar] [CrossRef]
- Capita, R.; Buzón-Durán, L.; Riesco-Peláez, F.; Alonso-Calleja, C. Effect of sub-lethal concentrations of biocides on the structural parameters and viability of the biofilms formed by Salmonella Typhimurium. Foodborne Pathog. Dis. 2017, 14, 350–356. [Google Scholar] [CrossRef]
- Ranieri, M.R.; Whitchurch, C.B.; Burrows, L.L. Mechanisms of biofilm stimulation by subinhibitory concentrations of antimicrobials. Curr. Opin. Microbiol. 2018, 45, 164–169. [Google Scholar] [CrossRef]
- Silveira, D.R.; da Rosa, J.V.; Kaefer, K.; Bach, L.G.; de Oliveira Barbosa, A.; Timm, C.D. Ability of Vibrio vulnificus isolated from fish of the Lagoa dos Patos estuary in south Brazil to form biofilms after sublethal stress and bacterial resistance to antibiotics and sanitizers. Int. J. Food Microbiol. 2019, 303, 19–25. [Google Scholar] [CrossRef]
- Shajahan, I.F.; Kandaswamy, D.; Lakshminarayanan, L.; Selvarajan, R. Substantivity of hypochlorous acid-based disinfectant against biofilm formation in the dental unit waterlines. J. Conserv. Dent. JCD 2017, 20, 2–5. [Google Scholar] [CrossRef]
- Iñiguez-Moreno, M.; Gutiérrez-Lomelí, M.; Guerrero-Medina, P.J.; Avila-Novoa, M.G. Biofilm formation by Staphylococcus aureus and Salmonella spp. under mono and dual-species conditions and their sensitivity to cetrimonium bromide, peracetic acid and sodium hypochlorite. Braz. J. Microbiol. 2018, 49, 310–319. [Google Scholar] [CrossRef]
- Capita, R.; Riesco-Peláez, F.; Alonso-Hernando, A.; Alonso-Calleja, C. Exposure of Escherichia coli ATCC 12806 to sublethal concentrations of food-grade biocides influences its ability to form biofilm, resistance to antimicrobials, and ultrastructure. Appl. Environ. Microbiol. 2014, 80, 1268–1280. [Google Scholar] [CrossRef] [Green Version]
- Devivilla, S.; Lekshmi, M.; Kumar, S.H.; Valappil, R.K.; Roy, S.D.; Nayak, B.B. Effect of Sodium Hypochlorite on Biofilm-Forming Ability of Histamine-Producing Bacteria Isolated from Fish. J. Food Prot. 2019, 82, 1417–1422. [Google Scholar] [CrossRef]
- Lin, H.; Zhu, X.; Wang, Y.; Yu, X. Effect of sodium hypochlorite on typical biofilms formed in drinking water distribution systems. J. Water Health 2017, 15, 218–227. [Google Scholar] [CrossRef] [Green Version]
- Toté, K.; Horemans, T.; Berghe, D.V.; Maes, L.; Cos, P. Inhibitory Effect of Biocides on the Viable Masses and Matrices of Staphylococcus aureus and Pseudomonas aeruginosa Biofilms. Appl. Environ. Microbiol. 2010, 76, 3135–3142. [Google Scholar] [CrossRef] [Green Version]
- Gomes, I.B.; Simões, M.; Simões, L.C. The effects of sodium hypochlorite against selected drinking water-isolated bacteria in planktonic and sessile states. Sci. Total Environ. 2016, 565, 40–48. [Google Scholar] [CrossRef] [Green Version]
- Mirani, Z.A.; Fatima, A.; Urooj, S.; Aziz, M.; Khan, M.N.; Abbas, T. Relationship of cell surface hydrophobicity with biofilm formation and growth rate: A study on Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli. Iran. J. Basic Med. Sci. 2018, 21, 760–769. [Google Scholar]
- Obe, T.; Nannapaneni, R.; Sharma, C.S.; Kiess, A. Homologous stress adaptation, antibiotic resistance, and biofilm forming ability of Salmonella enterica serovar Heidelberg ATCC8326 on different food-contact surfaces following exposure to sublethal chlorine concentrations1. Poult. Sci. 2018, 97, 951–961. [Google Scholar] [CrossRef]
- Fong, J.C.N.; Karplus, K.; Schoolnik, G.K.; Yildiz, F.H. Identification and Characterization of RbmA, a Novel Protein Required for the Development of Rugose Colony Morphology and Biofilm Structure in Vibrio cholerae. J. Bacteriol. 2006, 188, 1049–1059. [Google Scholar] [CrossRef] [Green Version]
- Bansal, M.; Nannapaneni, R.; Kode, D.; Chang, S.; Sharma, C.S.; McDaniel, C.; Kiess, A. Rugose Morphotype in Salmonella Typhimurium and Salmonella Heidelberg Induced by Sequential Exposure to Subinhibitory Sodium Hypochlorite Aids in Biofilm Tolerance to Lethal Sodium Hypochlorite on Polystyrene and Stainless Steel Surfaces. Front. Microbiol. 2019, 10, 2704–2720. [Google Scholar] [CrossRef] [Green Version]
- Strempel, N.; Nusser, M.; Neidig, A.; Brenner-Weiss, G.; Overhage, J. The Oxidative Stress Agent Hypochlorite Stimulates c-di-GMP Synthesis and Biofilm Formation in Pseudomonas aeruginosa. Front. Microbiol. 2017, 8, 2311–2325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schäper, S.; Krol, E.; Skotnicka, D.; Kaever, V.; Hilker, R.; Søgaard-Andersen, L.; Becker, A. Cyclic Di-GMP Regulates Multiple Cellular Functions in the Symbiotic Alphaproteobacterium Sinorhizobium meliloti. J. Bacteriol. 2016, 198, 521–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacey, M.M.; Partridge, J.D.; Green, J. Escherichia coli K-12 YfgF is an anaerobic cyclic di-GMP phosphodiesterase with roles in cell surface remodelling and the oxidative stress response. Microbiology 2010, 156, 2873–2886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenal, U.; Reinders, A.; Lori, C. Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 2017, 15, 271–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valentini, M.; Filloux, A. Biofilms and Cyclic di-GMP (c-di-GMP) Signaling: Lessons from Pseudomonas aeruginosa and Other Bacteria. J. Biol. Chem. 2016, 291, 12547–12555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hisert, K.B.; MacCoss, M.; Shiloh, M.U.; Darwin, K.H.; Singh, S.; Jones, R.A.; Ehrt, S.; Zhang, Z.; Gaffney, B.L.; Gandotra, S.; et al. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: Role of cyclic diGMP. Mol. Microbiol. 2005, 56, 1234–1245. [Google Scholar] [CrossRef]
- Farrant, K.V.; Spiga, L.; Davies, J.C.; Williams, H.D. Response of Pseudomonas aeruginosa to the innate immune system-derived oxidants hypochlorous acid and hypothiocyanous acid. bioRxiv 2020, in press. [Google Scholar] [CrossRef] [Green Version]
- Peeters, E.; Sass, A.; Mahenthiralingam, E.; Nelis, H.; Coenye, T. Transcriptional response of Burkholderia cenocepacia J2315 sessile cells to treatments with high doses of hydrogen peroxide and sodium hypochlorite. BMC Genom. 2010, 11, 90. [Google Scholar] [CrossRef] [Green Version]
- Giacomucci, S.; Cros, C.D.-N.; Perron, X.; Mathieu-Denoncourt, A.; Duperthuy, M. Flagella-dependent inhibition of biofilm formation by sub-inhibitory concentration of polymyxin B in Vibrio cholerae. PLoS ONE 2019, 14, e0221431. [Google Scholar] [CrossRef] [Green Version]
- Lipus, D.; Vikram, A.; Gulliver, D.; Bibby, K. Upregulation of peroxide scavenging enzymes and multidrug efflux proteins highlight an active sodium hypochlorite response in Pseudomonas fluorescens biofilms. Biofouling 2019, 35, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Lyu, M.; Wen, Y.; Song, Y.; Li, J.; Chen, Z. Organic Peroxide-Sensing Repressor OhrR Regulates Organic Hydroperoxide Stress Resistance and Avermectin Production in Streptomyces avermitilis. Front. Microbiol. 2018, 9, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-W.; Chung, C.-H.; Ma, T.-Y.; Wong, H. Roles of Alkyl Hydroperoxide Reductase Subunit C (AhpC) in Viable but Nonculturable Vibrio parahaemolyticus. Appl. Environ. Microbiol. 2013, 79, 3734–3743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinckx, T.; Wei, Q.; Matthijs, S.; Cornelis, P. The Pseudomonas aeruginosa oxidative stress regulator OxyR influences production of pyocyanin and rhamnolipids: Protective role of pyocyanin. Microbiology 2010, 156, 678–686. [Google Scholar] [CrossRef] [Green Version]
- Hou, A.-M.; Yang, D.; Miao, J.; Shi, D.-Y.; Yin, J.; Yang, Z.-W.; Shen, Z.-Q.; Wang, H.-R.; Qiu, Z.-G.; Liu, W.-L.; et al. Chlorine injury enhances antibiotic resistance in Pseudomonas aeruginosa through over expression of drug efflux pumps. Water Res. 2019, 156, 366–371. [Google Scholar]
- Zhang, Y.; Gu, A.Z.; He, M.; Li, D.; Chen, J. Subinhibitory Concentrations of Disinfectants Promote the Horizontal Transfer of Multidrug Resistance Genes within and across Genera. Environ. Sci. Technol. 2017, 51, 570–580. [Google Scholar] [CrossRef]
- Lin, H.; Ye, C.; Chen, S.; Zhang, S.; Yu, X. Viable but non-culturable E. coli induced by low level chlorination have higher persistence to antibiotics than their culturable counterparts. Environ. Pollut. 2017, 230, 242–249. [Google Scholar] [CrossRef]
- Nasr, A.M.; Mostafa, M.S.; Arnaout, H.H.; Elshimy, A.A.A. The effect of exposure to sub-inhibitory concentrations of hypochlorite and quaternary ammonium compounds on antimicrobial susceptibility of Pseudomonas aeruginosa. Am. J. Infect. Control 2018, 46, e57–e63. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Schweizer, H.P. Evidence of MexT-Independent Overexpression of MexEF-OprN Multidrug Efflux Pump of Pseudomonas aeruginosa in Presence of Metabolic Stress. PLoS ONE 2011, 6, e26520. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zeng, J.; Wang, Y.; Ye, C.; Zhu, S.; Feng, L.; Zhang, S.; Yu, X. Modelling the effect of chlorination/chloramination on induction of viable but non-culturable (VBNC) Escherichia coli. Environ. Technol. 2019. [Google Scholar] [CrossRef]
- Chen, S.; Li, X.; Wang, Y.; Zeng, J.; Ye, C.; Li, X.; Guo, L.; Zhang, S.; Yu, X. Induction of Escherichia coli into a VBNC state through chlorination/chloramination and differences in characteristics of the bacterium between states. Water Res. 2018, 142, 279–288. [Google Scholar] [PubMed]
- Ayrapetyan, M.; Williams, T.; Oliver, J. Relationship Between the Viable but Nonculturable State and Antibiotic Persister Cells. J. Bacteriol. 2018, 200, e00249-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fakruddin, M.; Mannan, K.S.B.; Andrews, S. Viable but Nonculturable Bacteria: Food Safety and Public Health Perspective. ISRN Microbiol. 2013, 2013, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhong, J.; Wei, C.; Lin, C.-W.; Ding, T. Current Perspectives on Viable but Non-culturable State in Foodborne Pathogens. Front. Microbiol. 2017, 8, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyström, T. Nonculturable bacteria: Programmed survival forms or cells at death’s door? BioEssays News Rev. Mol. Cell. Dev. Biol. 2003, 25, 204–211. [Google Scholar] [CrossRef]
- Oliver, J.D.; Dagher, M.; Linden, K. Induction of Escherichia coli and Salmonella typhimurium into the viable but nonculturable state following chlorination of wastewater. J. Water Health 2005, 3, 249–257. [Google Scholar] [CrossRef]
- Huang, K.; Zhang, X.-X.; Shi, P.; Wu, B.; Ren, H. A comprehensive insight into bacterial virulence in drinking water using 454 pyrosequencing and Illumina high-throughput sequencing. Ecotoxicol. Environ. Saf. 2014, 109, 15–21. [Google Scholar] [CrossRef]
- Lavoie, E.G.; Wangdi, T.; Kazmierczak, B.I. Innate immune responses to Pseudomonas aeruginosa infection. Microbes Infect. Inst. Pasteur 2011, 13, 1133–1145. [Google Scholar] [CrossRef] [Green Version]
- Coburn, B.; Sekirov, I.; Finlay, B.B. Type III Secretion Systems and Disease. Clin. Microbiol. Rev. 2007, 20, 535–549. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Marshall, N.C.; Rowland, J.L.; McCoy, J.M.; Worrall, L.J.; Santos, A.S.; Strynadka, N.C.J.; Finlay, B.B. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017, 15, 323–337. [Google Scholar] [CrossRef]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Cruz Nizer, W.S.; Inkovskiy, V.; Overhage, J. Surviving Reactive Chlorine Stress: Responses of Gram-Negative Bacteria to Hypochlorous Acid. Microorganisms 2020, 8, 1220. https://doi.org/10.3390/microorganisms8081220
da Cruz Nizer WS, Inkovskiy V, Overhage J. Surviving Reactive Chlorine Stress: Responses of Gram-Negative Bacteria to Hypochlorous Acid. Microorganisms. 2020; 8(8):1220. https://doi.org/10.3390/microorganisms8081220
Chicago/Turabian Styleda Cruz Nizer, Waleska Stephanie, Vasily Inkovskiy, and Joerg Overhage. 2020. "Surviving Reactive Chlorine Stress: Responses of Gram-Negative Bacteria to Hypochlorous Acid" Microorganisms 8, no. 8: 1220. https://doi.org/10.3390/microorganisms8081220