Effects of Weather and Environmental Factors on the Seasonal Prevalence of Foodborne Viruses in Irrigation Waters in Gyeonggi Province, Korea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Sites and Sample Collection
2.2. Virus Concentrations and RNA Extraction
2.3. Real-Time Reverse Transcription-Quantitative Polymerase Chain Reaction (Real-Time RT-qPCR)
2.4. Male-Specific Coliphage Isolation, Aerobic Plate Count (APC), and Coliform Count
2.5. Weather and Environmental Data
2.6. Statistical Analyses
3. Results
3.1. Detection of Viruses and Indicator Microorganisms
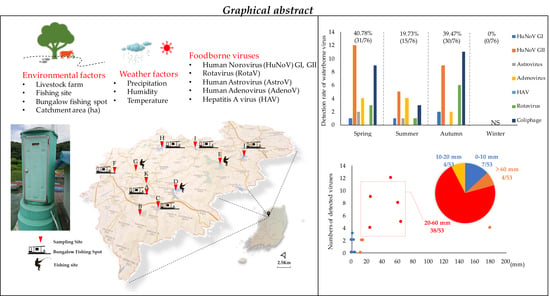
3.2. Seasonal Detection of Viruses and Indicator Microorganisms in Irrigation Water
3.3. Correlation between Detected Viruses and Weather Factors
3.4. Correlation between Detected Viruses and Environmental Factors
4. Discussion
4.1. Prevalence of Foodborne Viruses and Indicator Microorganisms in Reservoirs
4.2. Seasonal Prevalence of Foodborne Viruses in Reservoirs
4.3. Effects of Weather Factors on the Prevalence of Foodborne Viruses in Reservoirs
4.4. Effects of Environmental Factors on the Prevalence of Foodborne Viruses in Reservoirs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- IPCC. Global Warming of 1.5 °C. 2018. Available online: https://www.ipcc.ch/sr15/ (accessed on 28 July 2020).
- RAWRIS. Rural Agricultural Water Resource Information System. 2019. Available online: https://rawris.ekr.or.kr/awminfo/storagerate02.do (accessed on 28 July 2020).
- Kim, C.J.; Park, M.J.; Lee, J.H. Analysis of climate change impacts on the spatial and frequency patterns of drought using a potential drought hazard mapping approach. Int. J. Climatol. 2014, 34, 61–80. [Google Scholar] [CrossRef]
- Ko, D.-H. Drought Causes Water Shortage Crisis in Southern Korea. TheKoreaTimes (Labor & Environment). 2017. Available online: http://www.koreatimes.co.kr/www/nation/2017/07/371_231587.html (accessed on 28 July 2020).
- KMA. Climate of South Korea. 2019. Available online: https://www.weather.go.kr/w/index.do (accessed on 28 July 2020).
- Pall, E.; Niculae, M.; Kiss, T.; Sandru, C.D.; Spinu, M. Human impact on the microbiological water quality of the rivers. Food Environ. Virol. 2013, 62, 1635–1640. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Navratil, S.; Gregory, A.; Bauer, A.; Srinath, I.; Szonyi, B.; Nightingale, K.; Anciso, J.; Jun, M.; Han, D.; et al. Farm management, environment, and weather factors jointly affect the probability of spinach contamination by generic Escherichia coli at the preharvest stage. Appl. Environ. Microbiol. 2014, 80, 2504–2515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Navratil, S.; Gregory, A.; Bauer, A.; Srinath, I.; Szonyi, B.; Nightingale, K.; Anciso, J.; Jun, M.; Han, D.; et al. Multifactorial effects of ambient temperature, precipitation, farm management, and environmental factors determine the level of generic Escherichia coli contamination on preharvested spinach. Appl. Environ. Microbiol. 2015, 81, 2635–2650. [Google Scholar] [CrossRef] [Green Version]
- Iwu, C.D.; Okoh, A.I. Preharvest Transmission Routes of Fresh Produce Associated Bacterial Pathogens with Outbreak Potentials: A Review. Int. J. Environ. Res. Public Health 2019, 16, 4407. [Google Scholar] [CrossRef] [Green Version]
- Steele, M.; Odumeru, J. Irrigation water as source of foodborne pathogens on fruit and vegetables. J. Food Prot. 2004, 67, 2839–2849. [Google Scholar] [CrossRef]
- Cook, N.; Bertrand, I.; Gantzer, C.; Pinto, R.M.; Bosch, A. Persistence of hepatitis A virus in fresh produce and production environments, and the effect of disinfection procedures: A review. Food Environ. Virol. 2018, 10, 253–262. [Google Scholar] [CrossRef]
- Ibrahim, E.M.E.; El-Liethy, M.A.; Abia, A.L.K.; Hemdan, B.A.; Shaheen, M.N. Survival of E. coli O157:H7, Salmonella Typhimurium, HAdV2 and MNV-1 in river water under dark conditions and varying storage temperatures. Sci. Total Environ. 2019, 648, 1297–1304. [Google Scholar] [CrossRef]
- Pachepsky, Y.; Shelton, D.R.; McLain, J.E.; Patel, J.; Mandrell, R.E. Irrigation waters as a source of pathogenic microorganisms in produce: A review. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 2011; Volume 113, pp. 75–141. [Google Scholar]
- Hirneisen, K.A.; Kniel, K.E. Comparative uptake of enteric viruses into spinach and green onions. Food Environ. Virol. 2013, 5, 24–34. [Google Scholar] [CrossRef]
- Wei, J.; Jin, Y.; Sims, T.; Kniel, K.E. Internalization of murine norovirus 1 by Lactuca sativa during irrigation. Appl. Environ. Microbiol. 2011, 77, 2508–2512. [Google Scholar] [CrossRef]
- Yang, Z.; Chambers, H.; DiCaprio, E.; Gao, G.; Li, J. Internalization and dissemination of human norovirus and Tulane virus in fresh produce is plant dependent. Food Microbiol. 2018, 69, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Kokkinos, P.; Kozyra, I.; Lazic, S.; Soderberg, K.; Vasickova, P.; Bouwknegt, M.; Rutjes, S.; Willems, K.; Moloney, R.; de Roda Husman, A.M.; et al. Virological quality of irrigation water in leafy green vegetables and berry fruits production chains. Food Environ. Virol. 2017, 9, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Kniel, K.E. Pre-harvest viral contamination of crops originating from fecal matter. Food Environ. Virol. 2010, 2, 195–206. [Google Scholar] [CrossRef]
- Greening, G.E.; Cannon, J.L. Human and animal viruses in food (Including Taxonomy of Enteric Viruses). In Virus in Foods; Springer: Berlin/Heidelberg, Germany, 2016; pp. 5–57. [Google Scholar] [CrossRef]
- Kumazaki, M.; Usuku, S. Norovirus genotype distribution in outbreaks of acute gastroenteritis among children and older people: An 8-year study. BMC Infect. Dis. 2016, 16, 643. [Google Scholar] [CrossRef] [Green Version]
- Bosch, A.; Pintó, R.M.; Guix, S. Foodborne viruses. Curr. Opin. Food Sci. 2016, 8, 110–119. [Google Scholar] [CrossRef]
- Koopmans, M.; Duizer, E. Foodborne viruses: An emerging problem. Int. J. Food Microbiol. 2004, 90, 23–41. [Google Scholar] [CrossRef]
- Yugo, D.M.; Meng, X.-J. Hepatitis E virus: Foodborne, waterborne and zoonotic transmission. Int. J. Environ. Res. Public Health 2013, 10, 4507–4533. [Google Scholar] [CrossRef]
- Kim, S.H.; Cheon, D.S.; Kim, J.H.; Lee, D.H.; Jheong, W.H.; Heo, Y.J.; Chung, H.M.; Jee, Y.; Lee, J.S. Outbreaks of gastroenteritis that occurred during school excursions in Korea were associated with several waterborne strains of norovirus. J. Clin. Microbiol. 2005, 43, 4836–4839. [Google Scholar] [CrossRef] [Green Version]
- MFDS. Ministry of Food and Drug Safety (MFDS). 2018. Available online: https://www.foodsafetykorea.go.kr/portal/healthyfoodlife/foodPoisoningStat.do (accessed on 28 July 2020).
- Kim, J.S.; Lee, W.J.; Lee, S.K.; Lee, E.J.; Hyun, J.; Kim, H.S.; Song, W.; Kim, H.S. Molecular epidemiology of human astrovirus in stool samples from patients with acute gastroenteritis in Korea, 2013–2017. Ann. Lab. Med. 2019, 39, 367–372. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.I.; Song, M.O.; Chung, J.Y.; Han, T.H.; Ahn, Y.M.; Seo, J.W.; Kim, M.S.; Kim, M.Y.; Kim, W.Y.; Lee, C.H. Outbreak of rotavirus variant P (8) in Seoul, South Korea. J. Med. Virol. 2008, 80, 1661–1665. [Google Scholar] [CrossRef]
- Shin, E.; Kim, J.S.; Oh, K.H.; Oh, S.S.; Kwon, M.; Kim, S.; Park, J.; Kwak, H.S.; Chung, G.T.; Kim, C.J.; et al. A waterborne outbreak involving hepatitis A virus genotype IA at a residential facility in the Republic of Korea in 2015. J. Clin. Virol. 2017, 94, 63–66. [Google Scholar] [CrossRef]
- Lee, G.C.; Kim, M.J.; Nam, S.; Lee, C.H. Incidence and molecular characterization of hepatitis A viruses in Korean surface water between 2007 and 2010. Microbiol. Immunol. 2014, 58, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Joo, I.S.; Ju, S.Y.; Jeong, M.H.; Song, Y.H.; Kwak, H.S. Research on the contamination levels of norovirus in food facilities using groundwater in South Korea, 2015-2016. Food Environ. Virol. 2018, 280, 35–40. [Google Scholar] [CrossRef] [PubMed]
- USEPA. US EPA Office of Research and Development, Cincinnati, OH. ICR Microbial Laboratory Manual. EPA/600/R-95/178. 1996. Available online: https://www.epa.gov/ (accessed on 28 July 2020).
- Lee, H.; Kim, M.; Lee, J.E.; Lim, M.; Kim, M.; Kim, J.M.; Jheong, W.H.; Kim, J.; Ko, G. Investigation of norovirus occurrence in groundwater in metropolitan Seoul, Korea. Sci. Total Environ. 2011, 409, 2078–2084. [Google Scholar] [CrossRef] [PubMed]
- Conte, J.; Potoczniak, M.J.; Tobe, S.S. Using synthetic oligonucleotides as standards in probe-based qPCR. Biotechniques 2018, 64, 177–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajal, V.B.; McSwain, B.S.; Thompson, D.E.; Leutenegger, C.M.; Kildare, B.J.; Wuertz, S. Validation of hollow fiber ultrafiltration and real-time PCR using bacteriophage PP7 as surrogate for the quantification of viruses from water samples. Water Res. 2007, 41, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- ISO. Microbiology of the Food Chain—Horizontal Method for Determination of Hepatitis A Virus and Norovirus Using Real-Time RT-PCR—Part 1: Method for Quantification. ISO/TS 15216-1. 2017. Available online: https://www.iso.org/standard/65681.html (accessed on 28 July 2020).
- Kokkinos, P.; Kozyra, I.; Lazic, S.; Bouwknegt, M.; Rutjes, S.; Willems, K.; Moloney, R.; de Roda Husman, A.M.; Kaupke, A.; Legaki, E. Harmonised investigation of the occurrence of human enteric viruses in the leafy green vegetable supply chain in three European countries. Food Environ. Virol. 2012, 4, 179–191. [Google Scholar] [CrossRef]
- Jothikumar, N.; Cromeans, T.; Sobsey, M.; Robertson, B. Development and evaluation of a broadly reactive TaqMan assay for rapid detection of hepatitis A virus. Appl. Environ. Microbiol. 2005, 71, 3359–3363. [Google Scholar] [CrossRef] [Green Version]
- Freeman, M.M.; Kerin, T.; Hull, J.; McCaustland, K.; Gentsch, J. Enhancement of detection and quantification of rotavirus in stool using a modified real-time RT-PCR assay. J. Med. Virol. 2008, 80, 1489–1496. [Google Scholar] [CrossRef]
- Dai, Y.-C.; Xu, Q.-H.; Wu, X.-B.; Hu, G.-F.; Tang, Y.-L.; Li, J.-D.; Chen, Q.; Nie, J. Development of real-time and nested RT-PCR to detect astrovirus and one-year survey of astrovirus in Jiangmen City, China. Arch. Virol. 2010, 155, 977–982. [Google Scholar] [CrossRef]
- Lyman, W.H.; Walsh, J.F.; Kotch, J.B.; Weber, D.J.; Gunn, E.; Vinjé, J. Prospective study of etiologic agents of acute gastroenteritis outbreaks in child care centers. J. Pediatr. 2009, 154, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Seo, D.J.; Seo, J.; Oh, H.; Jeon, S.B.; Ha, S.-D.; Myoung, J.; Choi, I.-S.; Choi, C. Detection of viable murine norovirus using the plaque assay and propidium-monoazide-combined real-time reverse transcription-polymerase chain reaction. J. Virol. Methods 2015, 221, 57–61. [Google Scholar] [CrossRef]
- USEPA. Method 1601:Male-Specific (F+) and Somatic Coliphage in Water by Two-Step Enrichment Procedure; EPA: Washington, DC, USA, 2001; Volume 40.
- Schraft, H.; Watterworth, L. Enumeration of heterotrophs, fecal coliforms and Escherichia coli in water: Comparison of 3M™ Petrifilm™ plates with standard plating procedures. J. Microbiol. Methods 2005, 60, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Association, A.P.H.; Association, A.W.W. Standard Methods for the Examination of Water and Wastewate; Water Environment Federation: Alexandria, VA, USA, 1995. [Google Scholar]
- Hata, A.; Katayama, H.; Kojima, K.; Sano, S.; Kasuga, I.; Kitajima, M.; Furumai, H. Effects of rainfall events on the occurrence and detection efficiency of viruses in river water impacted by combined sewer overflows. Sci. Total Environ. 2014, 468, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Tolouei, S.; Burnet, J.B.; Autixier, L.; Taghipour, M.; Bonsteel, J.; Duy, S.V.; Sauve, S.; Prevost, M.; Dorner, S. Temporal variability of parasites, bacterial indicators, and wastewater micropollutants in a water resource recovery facility under various weather conditions. Water Res. 2019, 148, 446–458. [Google Scholar] [CrossRef] [PubMed]
- Cheong, S.; Lee, C.; Song, S.W.; Choi, W.C.; Lee, C.H.; Kim, S.J. Enteric viruses in raw vegetables and groundwater used for irrigation in South Korea. Appl. Environ. Microbiol. 2009, 75, 7745–7751. [Google Scholar] [CrossRef] [Green Version]
- Haramoto, E.; Kitajima, M.; Katayama, H.; Ohgaki, S. Real-time PCR detection of adenoviruses, polyomaviruses, and torque teno viruses in river water in Japan. Water Res. 2010, 44, 1747–1752. [Google Scholar] [CrossRef]
- Sartorius, B.; Andersson, Y.; Velicko, I.; De Jong, B.; Lofdahl, M.; Hedlund, K.O.; Allestam, G.; Wangsell, C.; Bergstedt, O.; Horal, P.; et al. Outbreak of norovirus in Vastra Gotaland associated with recreational activities at two lakes during August 2004. Scand. J. Infect. Dis. 2007, 39, 323–331. [Google Scholar] [CrossRef]
- Wyn-Jones, A.P.; Carducci, A.; Cook, N.; D’Agostino, M.; Divizia, M.; Fleischer, J.; Gantzer, C.; Gawler, A.; Girones, R.; Holler, C.; et al. Surveillance of adenoviruses and noroviruses in European recreational waters. Water Res. 2011, 45, 1025–1038. [Google Scholar] [CrossRef]
- Shin, H.; Park, H.; Seo, D.J.; Jung, S.; Yeo, D.; Wang, Z.; Park, K.H.; Choi, C. Foodborne viruses detected sporadically in the fresh produce and its production environment in South Korea. Foodborne Pathog. Dis. 2019, 16, 411–420. [Google Scholar] [CrossRef]
- De Giglio, O.; Caggiano, G.; Bagordo, F.; Barbuti, G.; Brigida, S.; Lugoli, F.; Grassi, T.; La Rosa, G.; Lucentini, L.; Uricchio, V.F.; et al. Enteric viruses and fecal bacteria indicators to assess groundwater quality and suitability for irrigation. Int. J. Environ. Res. Public Health 2017, 14, 588. [Google Scholar] [CrossRef] [PubMed]
- Mackowiak, M.; Leifels, M.; Hamza, I.A.; Jurzik, L.; Wingender, J. Distribution of Escherichia coli, coliphages and enteric viruses in water, epilithic biofilms and sediments of an urban river in Germany. Sci. Total Environ. 2018, 626, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Prez, V.E.; Martinez, L.C.; Victoria, M.; Giordano, M.O.; Masachessi, G.; Re, V.E.; Pavan, J.V.; Colina, R.; Barril, P.A.; Nates, S.V. Tracking enteric viruses in green vegetables from central Argentina: Potential association with viral contamination of irrigation waters. Sci. Total Environ. 2018, 637–638, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, M.N.F.; Elmahdy, E.M.; Chawla-Sarkar, M. Quantitative PCR-based identification of enteric viruses contaminating fresh produce and surface water used for irrigation in Egypt. Environ. Sci. Pollut. Res. 2019, 26, 21619–21628. [Google Scholar] [CrossRef] [PubMed]
- Silverman, A.I.; Akrong, M.O.; Amoah, P.; Drechsel, P.; Nelson, K.L. Quantification of human norovirus GII, human adenovirus, and fecal indicator organisms in wastewater used for irrigation in Accra, Ghana. J. Water Health 2013, 11, 473–488. [Google Scholar] [CrossRef]
- Verani, M.; Federigi, I.; Donzelli, G.; Cioni, L.; Carducci, A. Human adenoviruses as waterborne index pathogens and their use for Quantitative Microbial Risk Assessment. Sci. Total Environ. 2019, 651, 1469–1475. [Google Scholar] [CrossRef]
- De Serres, G.; Cromeans, T.L.; Levesque, B.; Brassard, N.; Barthe, C.; Dionne, M.; Prud’homme, H.; Paradis, D.; Shapiro, C.N.; Nainan, O.V.; et al. Molecular confirmation of hepatitis A virus from well water: Epidemiology and public health implications. J. Infect. Dis. 1999, 179, 37–43. [Google Scholar] [CrossRef]
- Lodder, W.J.; de Roda Husman, A.M. Presence of noroviruses and other enteric viruses in sewage and surface waters in The Netherlands. Appl. Environ. Microbiol. 2005, 71, 1453–1461. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.M.; Hsu, B.M.; Kao, P.M.; Chang, T.Y.; Hsu, T.K.; Ho, Y.N.; Yang, Y.C.; Huang, Y.L. Prevalence, quantification, and typing of human adenoviruses detected in river water in Taiwan. Environ. Sci. Pollut. Res. 2015, 22, 8359–8366. [Google Scholar] [CrossRef]
- Prevost, B.; Lucas, F.S.; Goncalves, A.; Richard, F.; Moulin, L.; Wurtzer, S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ. Int. 2015, 79, 42–50. [Google Scholar] [CrossRef]
- Haramoto, E.; Kitajima, M.; Hata, A.; Torrey, J.R.; Masago, Y.; Sano, D.; Katayama, H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018, 135, 168–186. [Google Scholar] [CrossRef] [PubMed]
- Kayed, D. Microbial Quality of Irrigation Water Used in the Production of Fresh Produce in Arizona. Ph.D. Thesis, University of Arizona, Tucson, AZ, USA, 2004. Available online: https://repository.arizona.edu/handle/10150/191270 (accessed on 26 July 2020).
- Seymour, I.J.; Appleton, H. Foodborne viruses and fresh produce. J. Appl. Microbiol. 2001, 91, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Fisman, D. Seasonality of viral infections: Mechanisms and unknowns. Clin. Microbiol. Infect. 2012, 18, 946–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farkas, K.; Cooper, D.M.; McDonald, J.E.; Malham, S.K.; de Rougemont, A.; Jones, D.L. Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Sci. Total Environ. 2018, 634, 1174–1183. [Google Scholar] [CrossRef]
- Ferguson, C.M.; Coote, B.G.; Ashbolt, N.J.; Stevenson, I.M. Relationships between indicators, pathogens and water quality in an estuarine system. Water Res. 1996, 30, 9. [Google Scholar] [CrossRef]
- Haramoto, E.; Katayama, H.; Oguma, K.; Koibuchi, Y.; Furumai, H.; Ohgaki, S. Effects of rainfall on the occurrence of human adenoviruses, total coliforms, and Escherichia coli in seawater. Water Sci. Technol. 2006, 54, 225–230. [Google Scholar] [CrossRef]
- Sassi, H.P.; van Ogtrop, F.; Morrison, C.M.; Zhou, K.; Duan, J.G.; Gerba, C.P. Sediment re-suspension as a potential mechanism for viral and bacterial contaminants. J. Environ. Sci. Health Part A-Toxic/Hazard. Subst. Environ. Eng. 2020, 1–8. [Google Scholar] [CrossRef]
- Pachepsky, Y.A.; Shelton, D.R. Escherichia Coli and fecal coliforms in freshwater and estuarine sediments. Crit. Rev. Environ. Sci. Technol. 2011, 41, 1067–1110. [Google Scholar] [CrossRef]
- McLain, J.E.; Williams, C.F. Seasonal variation in accurate identification of Escherichia coli within a constructed wetland receiving tertiary-treated municipal effluent. Water Res. 2008, 42, 4041–4048. [Google Scholar] [CrossRef]
- KMA. Climate of Korea. 2009. Available online: https://www.kma.go.kr/eng/biz/climate_01.jsp (accessed on 28 July 2020).
- Shamkhali Chenar, S.; Deng, Z. Environmental indicators for human norovirus outbreaks. Int. J. Environ. Res. Public Health 2017, 27, 40–51. [Google Scholar] [CrossRef]
- Guber, A.K.; Williams, D.M.; Quinn, A.C.D.; Tamrakar, S.B.; Porter, W.F.; Rose, J.B. Model of pathogen transmission between livestock and white-tailed deer in fragmented agricultural and forest landscapes. Environ. Model. Softw. 2016, 80, 185–200. [Google Scholar] [CrossRef]
- Oba, M.; Tsuchiaka, S.; Omatsu, T.; Katayama, Y.; Otomaru, K.; Hirata, T.; Aoki, H.; Murata, Y.; Makino, S.; Nagai, M. A new comprehensive method for detection of livestock-related pathogenic viruses using a target enrichment system. Biochem. Biophys. Res. Commun. 2018, 495, 1871–1877. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; De Keuckelaere, A.; Uyttendaele, M. Fate of foodborne viruses in the “Farm to Fork” chain of fresh produce. Compr. Rev. Food. Sci. Food Saf. 2015, 14, 755–770. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M. Tracing the sources of outbreaks of food-and waterborne viral disease and outbreak investigation using molecular methods. In Viruses in Food and Water; Elsevier: Amsterdam, The Netherlands, 2013; pp. 139–158. [Google Scholar]
- Givens, C.E.; Kolpin, D.W.; Borchardt, M.A.; Duris, J.W.; Moorman, T.B.; Spencer, S.K. Detection of hepatitis E virus and other livestock-related pathogens in Iowa streams. Sci. Total Environ. 2016, 566–567, 1042–1051. [Google Scholar] [CrossRef] [Green Version]
- Kavurmaci, M.; Apaydin, A. Assessment of irrigation water quality by a Geographic Information System–Multicriteria Decision Analysis-based model: A case study from Ankara, Turkey. Water Environ. Res. 2019, 91, 11. [Google Scholar] [CrossRef]
- Mushtaha, A.M.; Van Camp, M.; Walraevens, K. Quantification of recharge and runoff from rainfall using new GIS tool: Example of the Gaza Strip Aquifer. Water 2019, 11, 84. [Google Scholar] [CrossRef] [Green Version]




| Virus | Primer/Probe | Oligonucleotide Sequence | Amplification Conditions | Reference |
|---|---|---|---|---|
| HuNoV GI | QNIF4 | 5′-CGCTGGATGCGNTTCCAT-3′ | 95 °C 15 s, 60 °C 1 min (45 cycles) | [36] |
| NV1LCR | 5′-CCTTAGACGCCATCATCATTTAC-3′ | |||
| NVGG1p | 5′-FAM-TGGACAGGAGAYCGCRATCT–TAMRA-3′ | |||
| HuNoV GII | QNIF2 | 5′-ATGTTCAGRTGGATGAGRTTCTCWGA-3′ | 95 °C 15 s, 60 °C 1 min (45 cycles) | [36] |
| COG2R | 5′-TCGACGCCATCTTCATTCACA-3′ | |||
| QNIFs | 5′-FAM-AGCACGTGGGAGGGCGATCG-TAMRA-3′ | |||
| HAV | Forward P | 5′-GGTAGGCTACGGGTGAAAC-3′ | 95 °C 10 s, 55 °C 20 s (45 cycles) | [37] |
| Reverse P | 5′-AACAACTCACCAATATCCGC-3′ | |||
| HAV Probe | 5′-FAM-CTTAGGCTAATACTTCTATGAAGAGATGC-TAMRA-3′ | |||
| RotaV | MVP3-FDeg | 5′-ACCATCTWCACRTRACCCTC-3′ | 95 °C 15 s, 56 °C 1 min (45 cycles) | [38] |
| MVP3-R1 | 5′-GGTCACATAACGCCCCTATA-3′ | |||
| MVP3-Probe | 5′-FAM-ATGAGCACAATAGTTAAAAGCTAACACTGTCAA-TAMRA-3′ | |||
| AstroV | AstV F | 5′-CCDGCCAGRCTCACAGAAGAG-3′ | 94 °C 15 s, 55 °C 20 s (45 cycles) | [39] |
| AstV R | 5′-GACTTGCTAGCCATCACACTYC-3′ | |||
| AstV Probe | 5′-FAM-ACTCCATCGCATTTGGAGGGGAGGACC-TAMRA-3′ | |||
| AdenoV | JTVFF | 5′-AACTTTCTCTCTTAATAGACGCC-3′ | 95 °C 10 s, 55 °C 30 s, 72 °C 27 s (45 cycles) | [40] |
| JTVFR | 5′-AGGGGGCTAGAAAACAAAA-3′ | |||
| JTVFAP | 5′-FAM-CGAAGAGTGCCCGTGTCAGC-BHQ-3′ | |||
| MNV-1 | MNV 5036 | 5′-ACGCTCAGCAGTCTTTGTGA-3′ | 95 °C 15 s, 60 °C 45 s (45 cycles) | [41] |
| MNV 5088 | 5′-CTGGCCTCAGAGCCATTG-3′ | |||
| MNV 5060 | 5′-FAM-CGCTGCGCCATCACTCATCC-TAMRA-3′ |
| Reservoir | Longitude/Latitude | Catchment Area (ha) | Pondage (m3) | No. of Livestock Farms | No. of Bungalow Fishing Spots | No. of Fishing Sites |
|---|---|---|---|---|---|---|
| Site A | 36°59′03.0″ N 127°17′04.6″ E | 121 | 45 | 6 | 0 | 1 |
| Site B | 36°57′45.0″ N 127°16′56.8″ E | 60 | 3.5 | 0 | 0 | 0 |
| Site C | 36°58′14.4″ N 127°18′40.4″ E | 1240 | 4707 | 10 | 12 | 1 |
| Site D | 36°59′45.0″ N 127°19′54.5″ E | 4830 | 12,085 | 34 | 0 | 1 |
| Site E | 37°02′32.8″ N 127°24′26.3″ E | 225 | 992.4 | 0 | 0 | 1 |
| Site F | 37°01′08.3″ N 127°11′08.0″ E | 375 | 614 | 18 | 2 | 1 |
| Site G | 37°01′30.0″ N 127°14′28.1″ E | 105 | 39 | 3 | 0 | 1 |
| Site H | 37°04′57.6″ N 127°16′59.7″ E | 7100 | 15,217 | 170 | 105 | 4 |
| Site I | 37°04′36.0″ N 127°21′54.2″ E | 485 | 1859 | 3 | 9 | 1 |
| Site J | 37°03′27.3″ N 127°26′43.3″ E | 790 | 2932.4 | 7 | 14 | 2 |
| Site K | 37°00′08.0″ N 127°16′02.3″ E | 172,200 | - | - | - | - |
| Sampling Site | HuNoV GI | HuNoV GII | AstroV | AdenoV | HAV | RotaV | Detection Rate of Positive Samples | Indicator Microorganisms | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Male-Specific Coliphage | APC (log CFU/mL) | Coliform (log CFU/mL) | E. coli (CFU/mL) | ||||||||
| A | 0/9 | 2/9 | 0/9 | 1/9 | 0/9 | 0/9 | 2/9 (22.22%) | 1/9 | 2.86 ± 0.97 | 1.15 ± 0.75 | ND b |
| B | 0/9 | 0/9 | 0/9 | 0/9 | 0/9 | 0/9 | 0/9 (0.00%) | 0/9 | 3.33 ± 0.50 | 1.81 ± 0.26 | ND |
| C | 1/9 | 2/9 | 0/9 | 2/9 | 0/9 | 1/9 | 2/9 (22.22%) | 3/9 | 2.07 ± 0.82 | 1.44 ± 0.77 | ND |
| D | 0/9 | 1/9 | 0/9 | 0/9 | 0/9 | 0/9 | 1/9 (11.11%) | 0/9 | 2.63 ± 0.77 | 1.49 ± 0.65 | 1 ± 0 |
| E | 0/9 | 2/9 | 0/9 | 1/9 | 0/9 | 0/9 | 3/9 (33.33%) | 0/9 | 2.79 ± 0.67 | 1.95 ± 0.80 | ND |
| F | 0/9 | 2/9 | 0/9 | 1/9 | 0/9 | 0/9 | 3/9 (33.33%) | 1/9 | 3.14 ± 0.61 | 1.53 ± 0.80 | ND |
| G | 1/9 | 3/9 | 0/9 | 0/9 | 0/9 | 2/9 | 3/9 (33.33%) | 1/9 | 2.72 ± 0.60 | 1.84 ± 0.53 | 2.5 ± 2.12 |
| H | 0/9 | 3/9 | 0/9 | 0/9 | 0/9 | 2/9 | 4/9 (44.44%) | 2/9 | 2.50 ± 0.65 | 1.60 ± 0.63 | ND |
| I | 0/9 | 3/9 | 1/9 | 1/9 | 0/9 | 2/9 | 3/9 (33.33%) | 6/9 | 2.63 ± 0.51 | 1.69 ± 0.56 | 10 ± 0 |
| J | 0/9 | 3/9 | 0/9 | 0/9 | 0/9 | 1/9 | 3/9 (33.33%) | 4/9 | 2.32 ± 0.27 | 1.27 ± 0.60 | 1 ± 0 |
| K | 2/6 | 5/6 | 2/6 | 4/6 | 0/6 | 2/6 | 5/6 (83.33%) | 5/6 | 3.28 ± 0.29 | 2.10 ± 0.60 | 7.2 ± 7.98 |
| Total | 4/96 a | 26/96 | 3/96 | 10/96 | 0/96 | 10/96 | 29/96 | 23/96 | - | - | - |
| (4.16%) | (27.1%) | (3.12%) | (10.4%) | (0.00%) | (10.4%) | (30.2%) | (23.9%) | ||||
| Detection of Viruses and Male-Specific Coliphages | No. of Positive Samples | |
|---|---|---|
| Single | HuNoV GII | 6 |
| HuNoV GII + Male-specific coliphage | 5 | |
| AdenoV | 2 | |
| RotaV | 1 | |
| Double | HuNoV GII + RotaV | 2 |
| HuNoV GII + RotaV + Male-specific coliphage | 4 | |
| HuNoV GII + AdenoV + Male-specific coliphage | 2 | |
| Multiple | HuNoV GII + AdenoV + AstroV + Male-specific coliphage | 2 |
| HuNoV GII + AdenoV + RotaV + Male-specific coliphage | 1 | |
| HuNoV GI + HuNoV GII + RotaV + Male-specific coliphage | 1 | |
| HuNoV GI + HuNoV GII + AdenoV + Male-specific coliphage | 1 | |
| HuNoV GI + HuNoV GII + AdenoV + AstroV + Male-specific coliphage | 1 | |
| HuNoV GI + HuNoV GII + AdenoV + RotaV + Male-specific coliphage | 1 | |
| Total positive samples (positive rate) | 29 (30.2%) a | |
| Log10β (SE b) a | ||||||||
|---|---|---|---|---|---|---|---|---|
| Virus | Male-Specific Coliphages | HuNoV GII | AdenoV | RotaV | AstroV | HuNoV GI | HAV | |
| Weather factors | ||||||||
| Precipitation b | 0.714 (0.095) * | 0.713 (0.030) * | 0.706 (0.012) * | 0.642 (0.053) * | 0.511 (0.067) | 0.237 (0.044) | 0.452 (0.044) | ND c |
| Humidity | 0.366 (1.279) | 0.413 (0.388) | 0.428 (0.148) | 0.335 (0.651) | 0.072 (0.776) | −0.003 (0.449) | 0.331 (0.466) | ND |
| Temperature | −0.025 (0.315) | −0.002 (0.098) | 0.006 (0.038) | 0.412 (0.144) | −0.409 (0.163) | 0.248 (0.100) | −0.042 (0.113) | ND |
| Environmental factors | ||||||||
| Catchment area | 0.233 (0.139) | 0.263 (0.143) | 0.296 (0.092) | −0.130 (0.094) | 0.281 (0.110) | −0.009 (0.049) | −0.110 (0.065) | ND |
| Pondage | 0.432 (0.076) | 0.385 (0.081) | 0.489 (0.050) | 0.107 (0.055) | 0.296 (0.065) | 0.130 (0.028) | −0.090 (0.038) | ND |
| Livestock | 0.267 (0.140) | 0.027 (0.145) | 0.356 (0.092) | −0.180 (0.095) | 0.281 (0.112) | −0.157 (0.049) | −0.065 (0.066) | ND |
| Bungalow fishing spot | 0.530 (0.117) | 0.737 (0.097) * | 0.307 (0.080) | 0.002 (0.091) | 0.663 (0.083) | 0.207 (0.046) | −0.016 (0.630) | ND |
| Fishing site | 0.605 (0.451) | 0.480 (0.518) | 0.745 (0.255) * | −0.159 (0.371) | 0.535 (0.384) | −0.055 (0.193) | −0.083 (0.257) | ND |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Shin, H.; Jung, S.; Yeo, D.; Park, H.; Shin, S.; Seo, D.J.; Park, K.H.; Choi, C. Effects of Weather and Environmental Factors on the Seasonal Prevalence of Foodborne Viruses in Irrigation Waters in Gyeonggi Province, Korea. Microorganisms 2020, 8, 1224. https://doi.org/10.3390/microorganisms8081224
Wang Z, Shin H, Jung S, Yeo D, Park H, Shin S, Seo DJ, Park KH, Choi C. Effects of Weather and Environmental Factors on the Seasonal Prevalence of Foodborne Viruses in Irrigation Waters in Gyeonggi Province, Korea. Microorganisms. 2020; 8(8):1224. https://doi.org/10.3390/microorganisms8081224
Chicago/Turabian StyleWang, Zhaoqi, Hansaem Shin, Soontag Jung, Daseul Yeo, Hyunkyung Park, Sangah Shin, Dong Joo Seo, Ki Hwan Park, and Changsun Choi. 2020. "Effects of Weather and Environmental Factors on the Seasonal Prevalence of Foodborne Viruses in Irrigation Waters in Gyeonggi Province, Korea" Microorganisms 8, no. 8: 1224. https://doi.org/10.3390/microorganisms8081224